Install tutorial
Install R

R is available for free for Windows, GNU/Linux and MacOS.
As for now, the latest version is 4.1.0.
One common complain about R is the frequency of updates. This is true, but can be sorted out with a good package manager. Of note, major updates are the only one forcing you to re-install extra-packages and are released on a yearly basis.
Windows
Visit the download page and choose the corresponding installer for your platform. Mind that you will need Rtools of the same version as your R to compile and install some packages.
MacOS
Visit the download page and choose the corresponding installer for your platform. You must have macOS 10.13 (High Sierra) or higher.
GNU/Linux
Any GNU/Linux distributions is bundled with a great package manager such as dpkg or rpm. Visit the appropriate web-page for your distribution here.
After you get the correct entry in your package manager, it should be quite easy as:
sudo apt-get install r-base r-base-devInstall Rstudio

Rstudio is an Integrated Development Editor, it wraps and interfaces R but, R needs to be installed first. The free-version contains everything you need.
Visit the download page and choose the corresponding installer for your platform. As for now, the latest version is 1.4.1106.
See the blog entry about this major release.
Open rstudio
They are 4 main panels in rstudio, but as the top-left is for scripting and by default missing, the layout should looks like
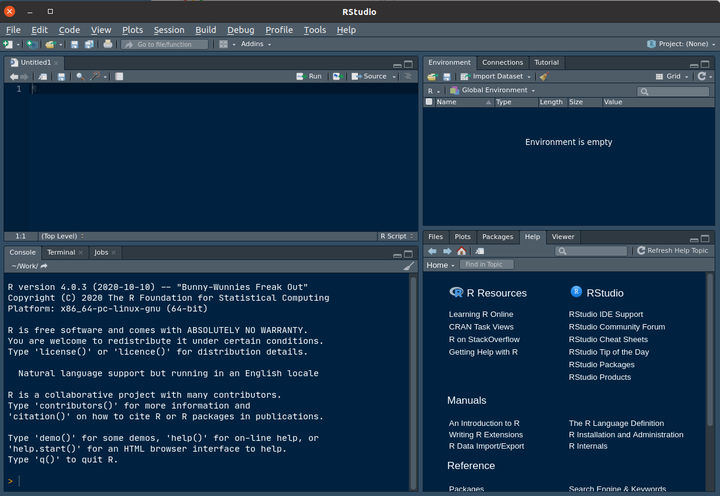
Of note, the default theme is with a white background. You can change it in the Global options > Appearance > Editor theme. Here is displayed the Cobalt theme.
install R packages from the Comprehensive R Archive Network (CRAN)
On the bottom-right panel, 5 tabs are present:
- Files
- Plots
- Packages
- Help
- Viewer
Click on the Packages tab (1.) and select the select the Install button (2.). Type tidyverse, remotes in (3.)
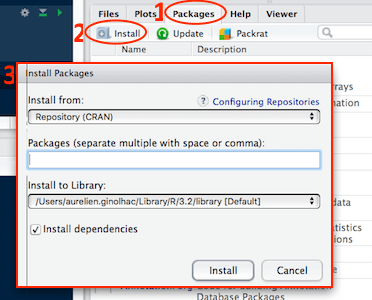
It takes some time, usually 10 seconds up to 5 minutes for a single package depending on its size and compilation stage.
The packages can also be installed in the console (i.e. the bottom-left panel). The snapshot below shows you how to install remotes using the console:
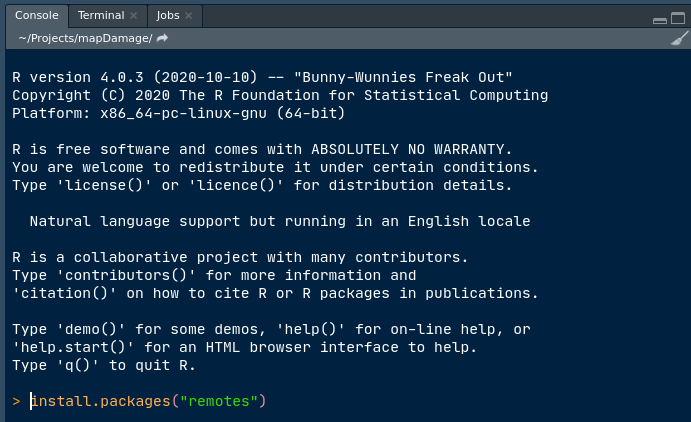
Of note, remotes is handy when you want to install a development version, for example if a bug was fixed but the package not yet submitted to CRAN.
remotes::install_github("r-lib/remotes")A note about package updates
The complaint of very frequent updates might be legitimate as it could be cumbersome to maintain R up-to-date when you have many packages. Actually, it is as easy as:
update.packages()But, you could also use the green Update button next to the Install (located in the Packages tab). The advantage is that you can check which package you would like to update and look at the actual changes by clicking on the NEWS link.
Installing bioconductor packages
The bioconductor resource will be detailed later, but to save time, copy / paste the following code into the console to install for exmple the GEOquery package:
# to install the version 3.8 of the bioconductor installer.
install.packages("BiocManager")
BiocManager::install("GEOquery")Package updates
There is no interface for bioconductor packages in Rstudio thus you need to run the following lines to update them (you will be asked if you want to update some / all / none):
BiocManager::install()Install the tidyverse
Several libraries are mandatory for successfully installing the tidyverse. If you encountered issues, install them with:
- in a terminal, for GNU/Linux:
sudo apt install -y libcurl4-openssl-dev libssl-dev libxml2-dev- in a terminal, for MacOS and
brew:
brew install libxml2 opensslthen in R, install the tidyverse.
install.packages("tidyverse")If you several cores, you may speedup the install with multi-threading (here for 2 cores):
install.packages("tidyverse", Ncpus = 2L)Testing your installation
Copy / paste the code below and you should obtain the following plot.
library(tidyverse)## Warning in system("timedatectl", intern = TRUE): running command 'timedatectl'
## had status 1mtcars %>%
pivot_longer(cols = c(drat, wt),
names_to = "measurement",
values_to = "value") %>%
ggplot(aes(x = value, y = mpg, colour = measurement)) +
geom_point() +
geom_smooth(method = "loess") +
theme_bw(14)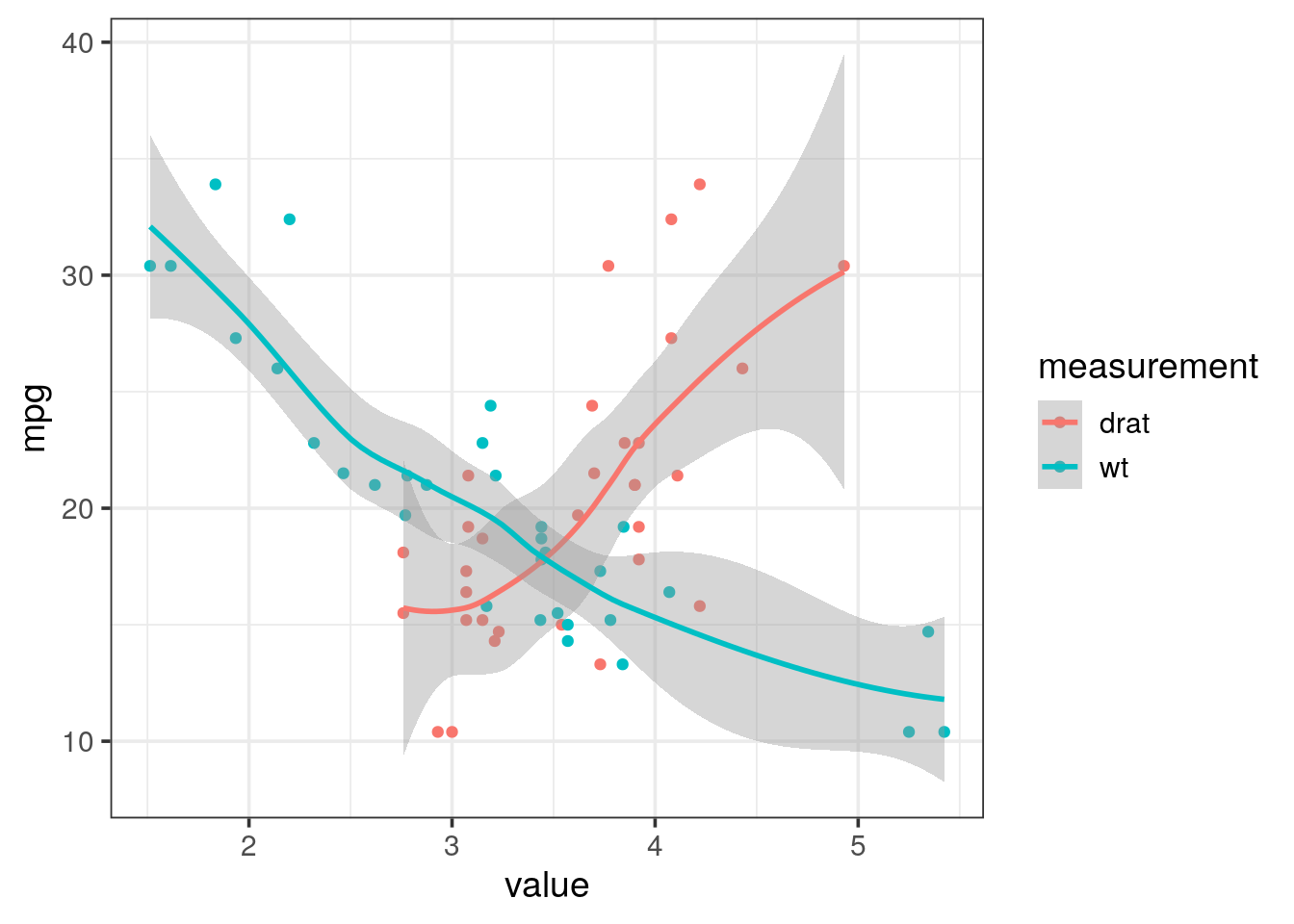
If not, please contact me with the error produced
