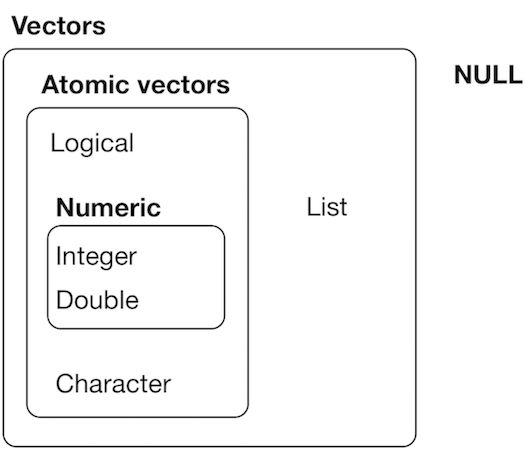
4 main types
| Type | Example |
|---|---|
| numeric | integer (2), double (2.34) |
| character (strings) | "tidyverse!" |
| boolean | TRUE / FALSE (T/F not protected) |
| complex | 2+0i |
Missing data and special cases
NA # not available, missing data
NA_real_
NA_integer_
NA_character_
NA_complex_
NULL # empty
-Inf/Inf # infinite values
NaN # Not a Number2L[1] 2typeof(2L)[1] "integer"2.34[1] 2.34typeof(2.34)[1] "double""tidyverse!"[1] "tidyverse!"TRUE[1] TRUE2+0i[1] 2+0i