setwd("C:/Users/veronica.codoni/Projects/Survival_Analysis")Setting up your work environment
Development tools, version control, packages
R Workshop
Monday, 16 September 2024
Introduction
Motivation
We want to work in a minimally distraction environment.
We want to write as little code as possible and use ideas that others have implemented.
How can we share code when it’s constantly changing?
Learning objectives


Integrated development environment
- Get to know RStudio the most important IDE for R projects
- Recommended and hidden features
- Use a versioning system
- Good package management
Project environment
- Set up a new scientific analysis project
- Set up code versioning with
git - Very basic use of
git - Set up and basic use of
renvfor environment management
RStudio
What is RStudio?
RStudio is an Integrated Development Environment.

Install R first
RStudio and R are independent.
Features
- Projects to ease files organisation
- Console to run R, with syntax highlighter
- full support for Rmarkdown docs & chunks
- Viewer for data / plots / website
- Package management (including building, tests and development)
- Autocompletion using TAB
- Cheatsheets
- Git integration for versioning
- Inline outputs
- Keyboard shortcuts
- integrated Terminal
- Jobs for running long runs in a separated session
The four panels layout
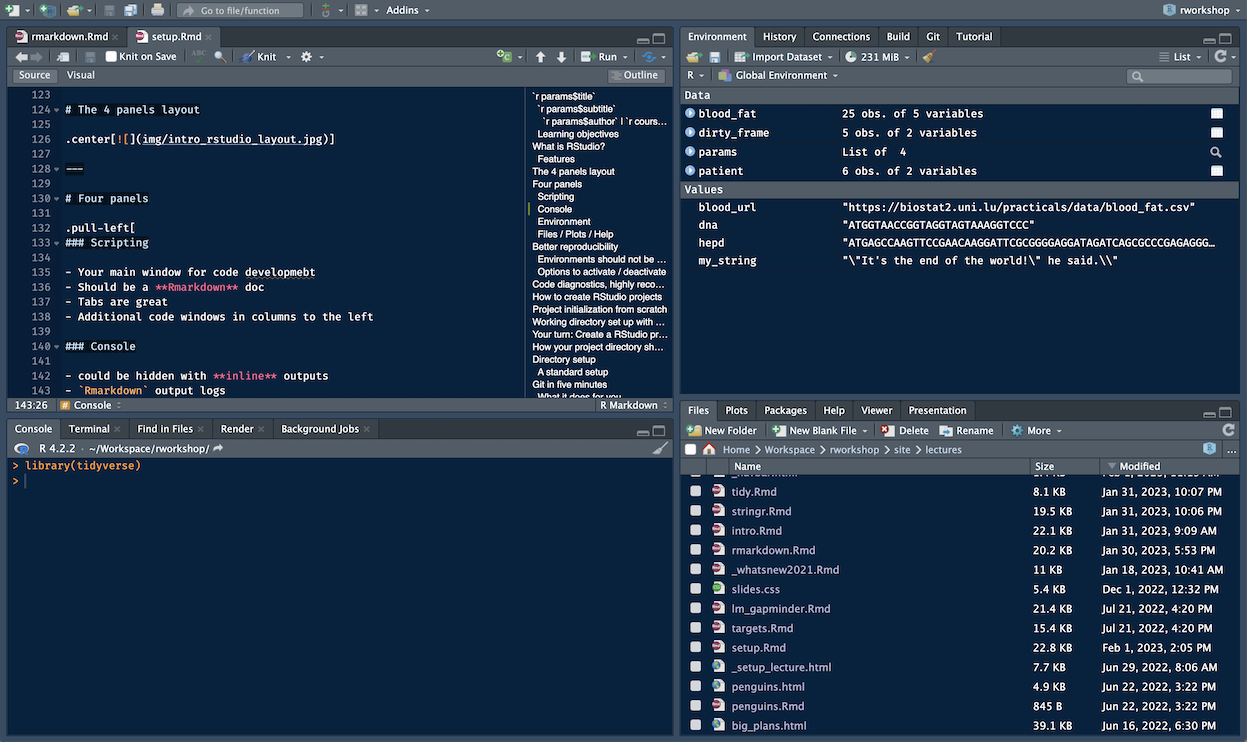
Four panels
Scripting
- Your main window for code development
- Should be a Rmarkdown doc
- Tabs are great
- Additional code windows in columns to the left
Console
- could be hidden with inline outputs
Rmarkdownoutput logs- optional, embed a nice
Terminaltab - optional,
Jobstab
Environment
- Environment, display loaded objects and their
str() - History is useless IMO
- nice
gitintegration - Build for development
- database connections interface
- Tutorial integration of
learnr
Files / Plots / Help
- Packages management tab
- unnecessary Plots panel when using inline outputs
- Help tab
- Viewer for animations
Better reproducibility
Environments should not be saved
rm(list = ls()) is not recommended
- does not make a fresh R session
library()calls remain- working directory not set!
- modified functions, evil
==<-!=
- Knitting
Rmarkdownfiles solves it- Always run in a fresh session
- Workflows like
targetsallow to be fast and fully reproducible
Options to activate / deactivate
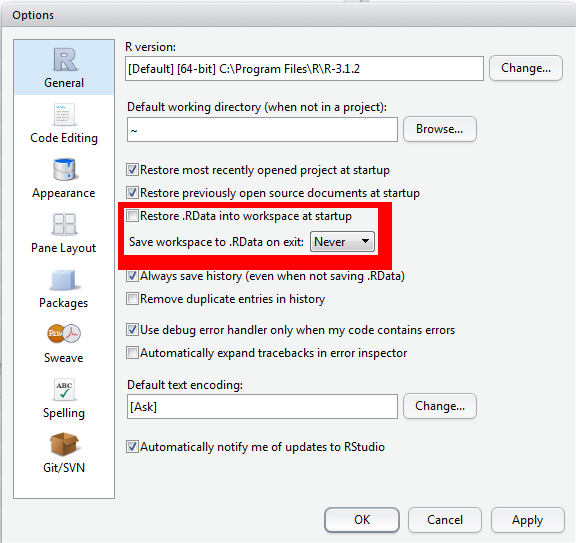
Warning
Please save all environments except for the R session
Other settings of note
Base pipe with Ctrl/⌘-Shift-M
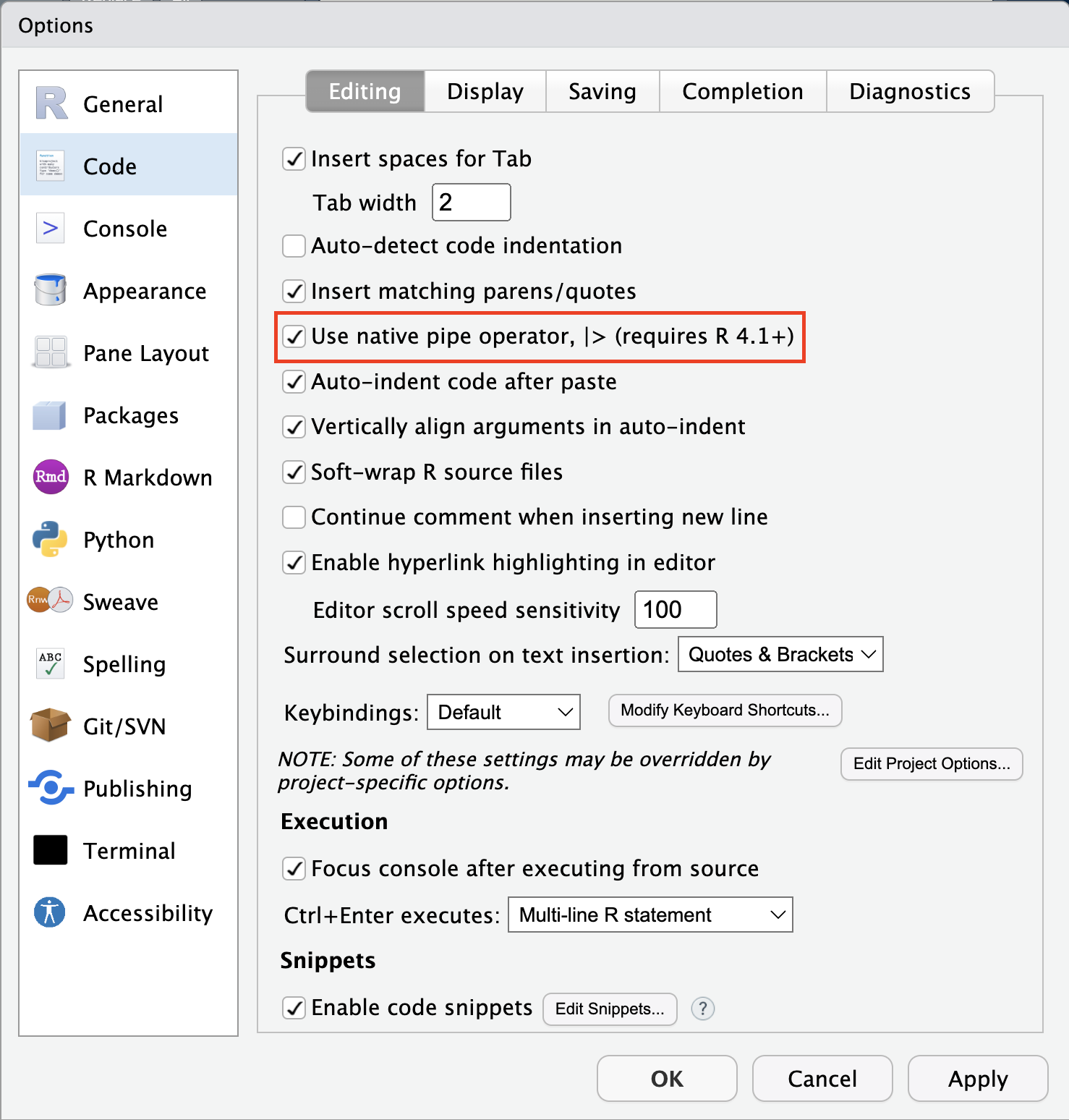
Additional code diagnostics
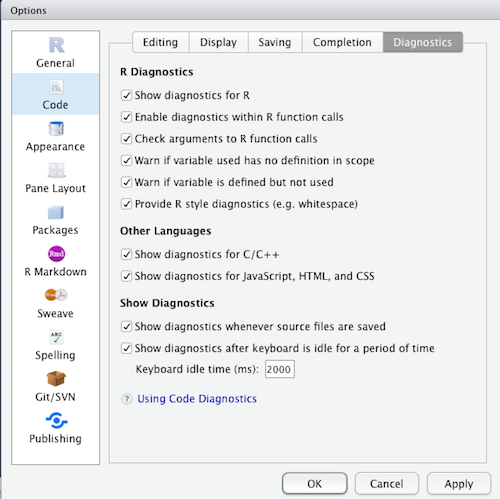
Consider at your own risk
Creates many warnings if all options are selected, particular with tidyverse code using non-standard evaluation.
Projects
RStudio projects
Set your working directory
- Any coding, including testing, you conduct should happen in the context of a project.
- A project in RStudio consists of a directory/folder containing a file with an
.Rprojextension. - The directory that contains this file is the reference for all paths in your project.
- A good project name goes a long way.
- Make it unique!
- There are multiple ways to create projects
- RStudio User Interface
- Place an
.Rprojfile in a already existing folder - Copy (clone) an existing setup under a different name
RStudio projects
Creating a project
- Use the Project button in the top right corner
- Select
New project
- Name the project properly
- A
.Rprojextension file is generated.
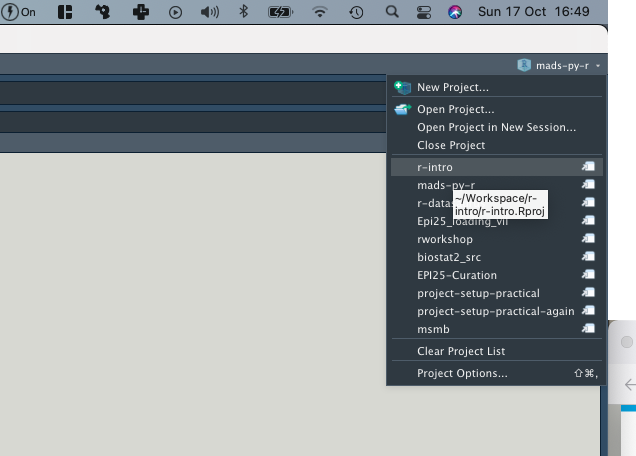
Project initialization from scratch
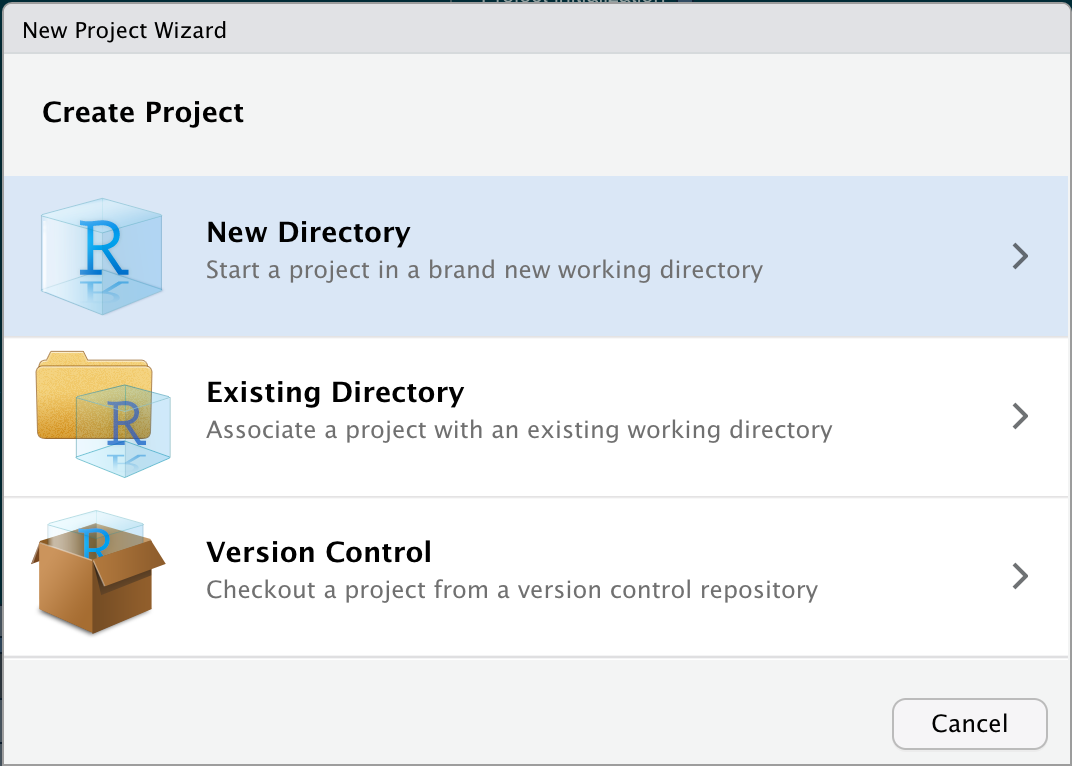
Good practice
Use git and renv when starting projects from scratch.
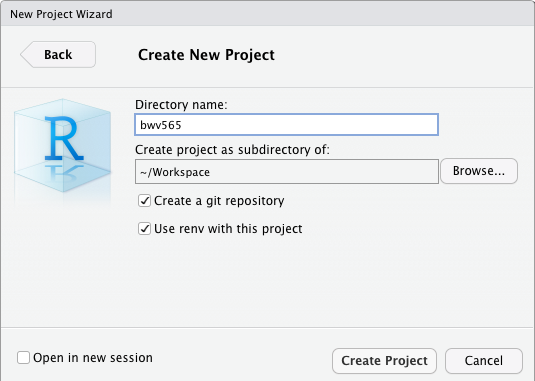
Working directory set up with RStudio projects
Avoid using setwd() and absolute file paths
What’s wrong with setwd()?1
- setwd() set the working directory to a specific folder through an absolute file path
- What if this folder moved to /Downloads, or onto another machine?
This approach is neither self-contained nor portable!
RStudio projects as alternative
Start a new research project/data analysis by creating a new Rstudio Project
- it helps to keep all the files together
- it sets automatically the working directory to the project directory
- it helps resolve file path issues by using relative paths
Create a Project in Rstudio assures the project directory to be stand-alone and portable.
Your turn: Create a RStudio project
Use the Project button in the top right corner.
Always name the projects properly
Keeping names concise, no spaces
Once the project has been created, a
.Rprojextension file is generated. This allows for automatic working directory set-up.
Exercise
- You can create a new project called
dummy-project- in a new folder
- enable
git - enable
renv
- Create sub-folders
dataandR. - Standard practice is to include an
R/project for code, similar tosrcandbindirectories.
Learn to switch between projects
We will not use this directory in this course any more, hence the name.
How your project directory should not look like
├── analysis.R
├── analusis_2.R
├── analusis4.R
├── parse_data.sh
├── tools # <- environment
│ ├── FACS.exe
│ ├── plink1.09
│ ├── plink1.7
│ ├── plink2.0
│ └── R2.9
├── mydata # <- see next lecture
│ ├── facs.xlsx
│ └── single-cells.txt
├── old_scripts/
│ ├── analysis_final.R
│ ├── analysis_final_2.R
│ ├── analysis_tmp.R
│ └── analysis_final_nature.R
├── Analysis-international_frontiers_biomedical-Machine_learning.R # IF 3.011!
├── Manuscript_2015-oct.docx
├── Manuscript_15-oct2.docx
├── Manuscript_15-oct2-PI.docx
└── Manuscript_Sept-2015.docxDirectory setup
A standard setup
Keeping things tidy
- How to keep older versions of scripts?
- How not to lose good, working code when changing things?
- How to share code with others for collaboration?
Version control
Code versioning systems support you in all of these use cases.
Code versioning
Git in five minutes

What it does for you
- Version your code at small scale
- No more physical copies of source code(!)
- Go back to the versions that worked
- Go back to the version you used a year ago
- Helps to document your advancements and solutions
- Share and publish your code while continuing to work on it
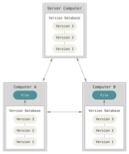
Capacities in code versioning
- Register (
add) files in a project called arepository - Track changes (
commit) resolutions.) - Exchange code with others through a
shared repository - The most common platforms for sharing repositories are
- GitHub, a global platform owned and supported by Microsoft
gitlab, local platforms maintained and supported by institutes, e.g. gitlab.lcsb.uni.lu- Both platforms offer many other tools
- Here, we cover Git as far as easily accessible from RStudio and other IDEs
Project initialization from a repository

From an existing Git repository
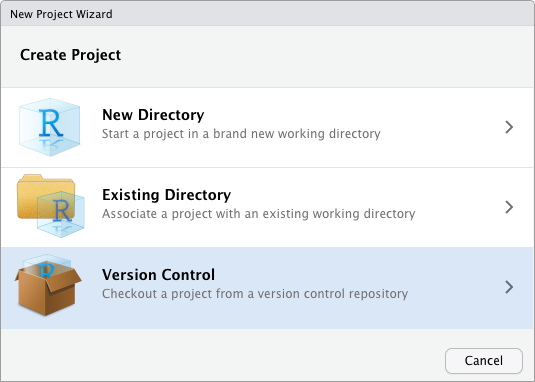
From a web repository by cloning (SSH)
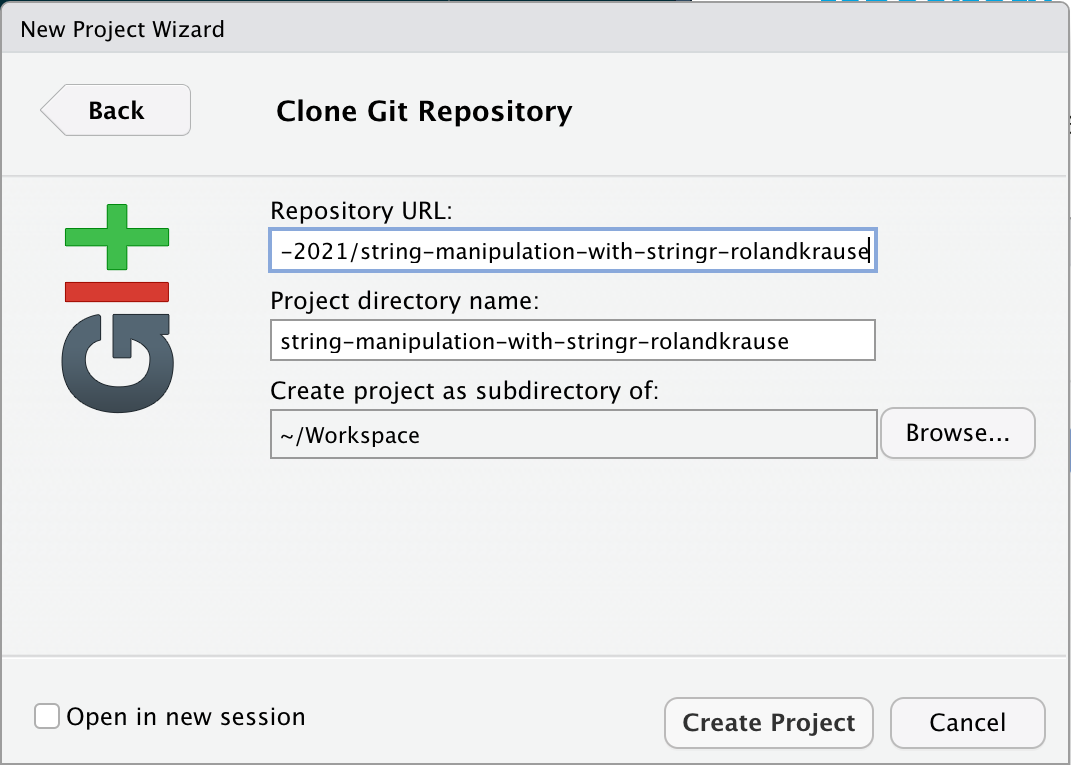
Working with the repository

Add a file to the staging area
Using RStudio git panel:
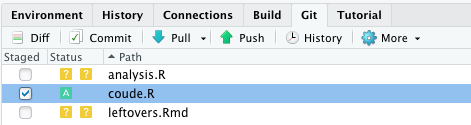
The equivalent command line is git add coude.R
Commit (to) changes

After a change file is tagged with M (modified)
Click in the staged column to move the modification to the staged area.
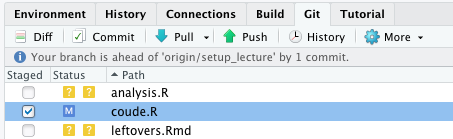
- Changes you do not commit are not tracked.
- In
gityou can move between commits. - All these changes are local unless you declare a remote.
The command line equivalent is:
git commit -m "Added Chip-seq input analysis.R"
Git history
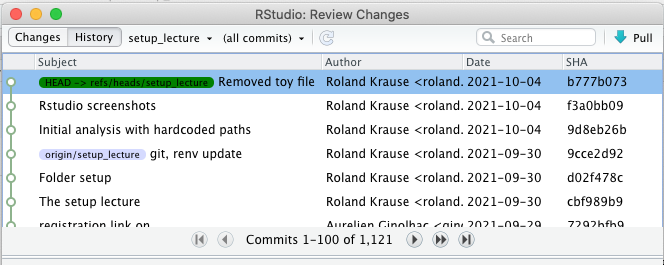
Synchronizing with a repository

Optionally use rebase (rewrite for linear history to avoid merge commits) using the little black arrow menu next to Pull
- Compare all files already in the remote repository
- Fail if changes are not committed
- If changes are committed.
- If additional changes have happened, ensure that code is working
Command line equivalents
- :
git pull - Pull with rebase:
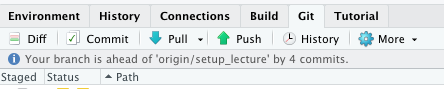
git pull --rebase Pushthe local state to the remote repository
Command line equivalent of :
git push
What not to put into Git

Data types to be aware of
- Sensitive data (should not be on GitHub)
- Large data
- Git servers are not setup to handle large files (>1Mb)
- Binary data
- git makes line by line comparisons
- Derived data
- Any data you can reproduce by computation
- You might want to archive some intermediate files by other means
Possible solutions
- Install scripts
- Copy or link your data to the
datadirectory
- Copy or link your data to the
- git-lfs Large File Storage
- Put files on server but only tracks via checksums
- Repository only tracks a small text file instead
Make your life easy with .gitignore

Working with Git

Helpful tips
1. Commit and pull frequently
- Minimally once daily when your code is working
- When adding a feature or analysis step (e.g. chunk-wise)
- Regularly merge from other branches if you have a multi-branch setup with others
2. Best learn from experienced person
- There is more than one productive set up to working with Git
- It’s a habit that needs developing
- You are the first to benefit from it
- You need to version files. Automate it. Don’t reinvent the wheel
Environment management
Packages and other programs
Packages

Dedicated to biology research {limma} example
Requires dedicated package BiocManager for installation.
Typical install
Package installation
CRAN install from Rstudio (autocompletion)
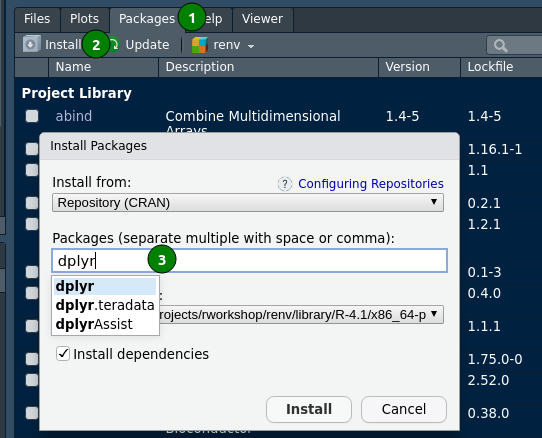
CRAN install from Rstudio’s console
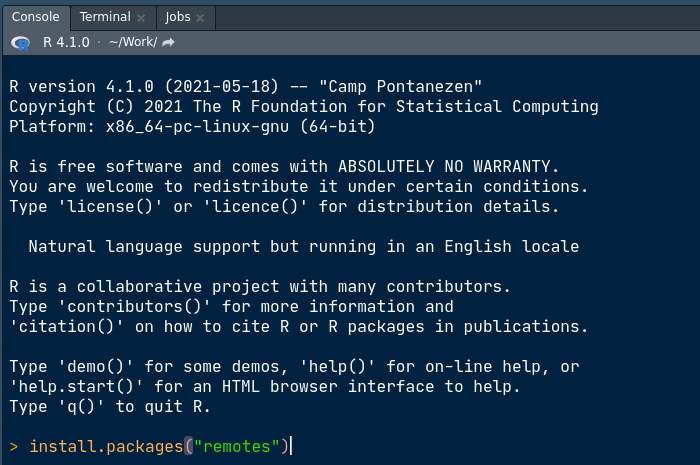
Managing your programming environment by project
Project documentation
- Source and version of package, incl. dependencies
- R/Bioconductor or Python versions
- (Ideally your work should not depend on the operating system)
Package documentations becomes complex fast and tedious to control manually * Projects require different versions of packages * Collaborators have different versions installed on their machines!
What does not work
- Agreements on specific versions in the group/institute/collaboration
- Hope that the operating systems/programming language will be supported until things are finished
- Keeping things as they are and not upgrading until the paper/revision/is out
- … not upgrading until the Internship is finished
- … not upgrading until the Master, PhD, Postdoc is finished
Packages for environment definition
condafor scientific programmingPython virtual environments
- Defines Python version and packages
renvR - environments- Defines packages from CRAN, Bioconductor, GitHub repositories
- Defines specific version (incl. R if you want)
- Every project has it’s own and defined packages
- Simplifies (!) installation
renv features
Basic functions
init()intializes your project and searches source code files for library callsdependencies()reads your calls
install(pkgs)installs packagepkgsincluding its dependenciessnapshot()registers changes, hashes and originrestore()to a certain point in time

Source: renv Vignette
Snapshot
Snapshot
```{r}
> renv::snapshot()
The following package(s) will be updated in the lockfile:
# CRAN ===============================
- RcppParallel [5.0.2 -> 5.0.3]
- cli [2.3.0 -> 2.3.1]
- pkgload [1.1.0 -> 1.2.0]
- tint [0.1.3 -> *]
# GitHub =============================
- targets [ropensci/targets@main: 598d7a23 -> bdc1b29c]
Do you want to proceed? [y/N]:
```renv.lock file after a snapshot
"R": {
"Version": "4.0.3",
"Repositories": [
{
"Name": "CRAN",
"URL": "https://cloud.r-project.org"
}
]
},
"Bioconductor": {
"Version": "3.12"
},
"Packages": {
"AnnotationDbi": {
"Package": "AnnotationDbi",
"Version": "1.52.0",
"Source": "Bioconductor",
"Hash": "ca5106b296b3aa6af713ce197be547c1"
},
"BH": {
"Package": "BH",
"Version": "1.75.0-0",
"Source": "Repository",
"Repository": "CRAN",
"Hash": "e4c04affc2cac20c8fec18385cd14691"
},
"targets": {
"Package": "targets",
"Version": "0.1.0.9000",
"Source": "GitHub",
"RemoteType": "github",
"RemoteUsername": "ropensci",
"RemoteRepo": "targets",
"RemoteRef": "main",
"RemoteSha": "598d7a23661d4c760209c7991bf10584eadcf7c8",
"RemoteHost": "api.github.com",
"Hash": "ee66061fd5c757ec600071965d457818"
},
[...]Sharing renv lock files
- Call
renv::init() - Add and commit
renv.lock.Rprofilerenv/settings.jsonrenv/activate.R
GitHub classroom configuration
Installation
Complete the install tutorial if you haven’t already including the Github setup with SSH
Make sure that your Github account is configured with SSH.
Check the github-classroom
Follow this link and authorize it
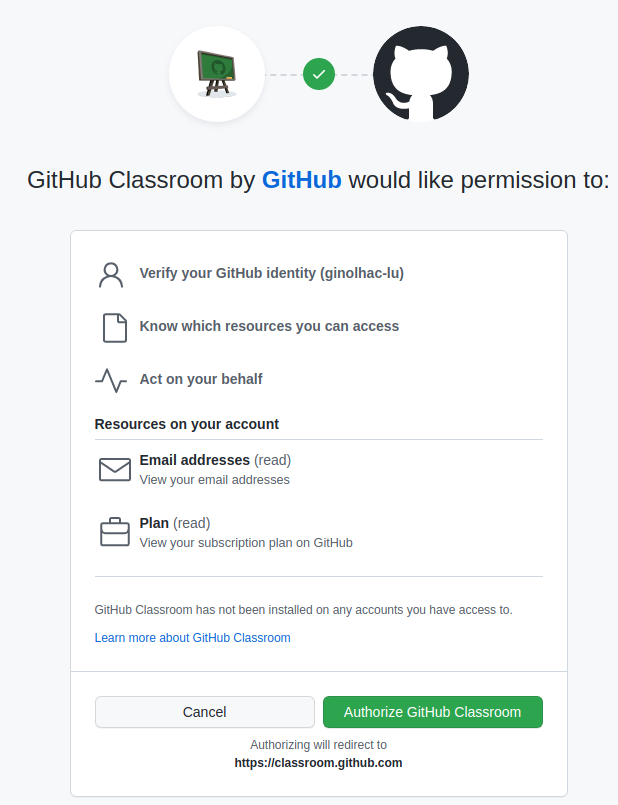
2. Accept the assignment
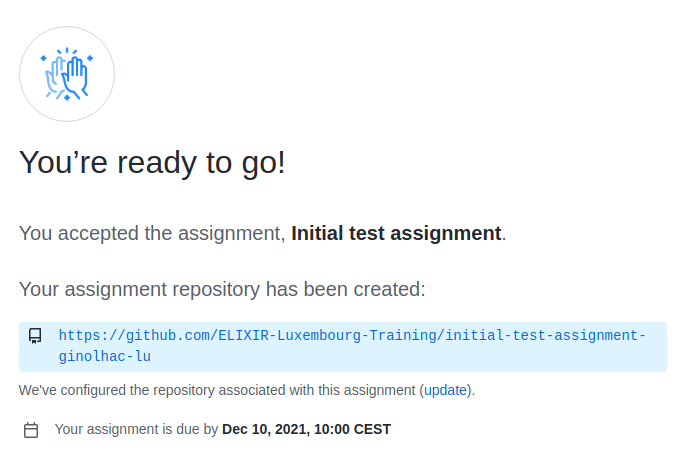
You should see this invite:
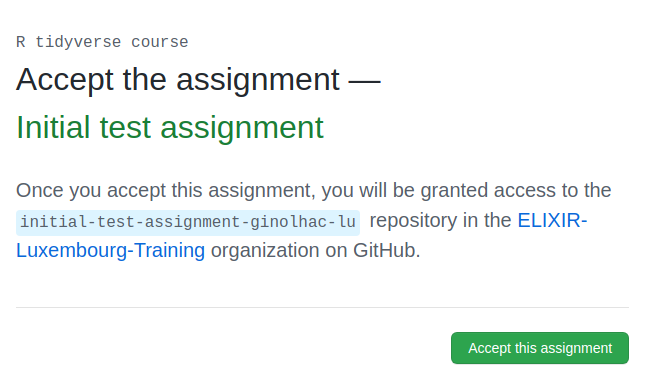
Check the repository
Go to the assignment
Reload the browser page
The assignment is created in your personal github repository.
![]()
Click on the link with the blue background
Check your own repo
It should look like this:
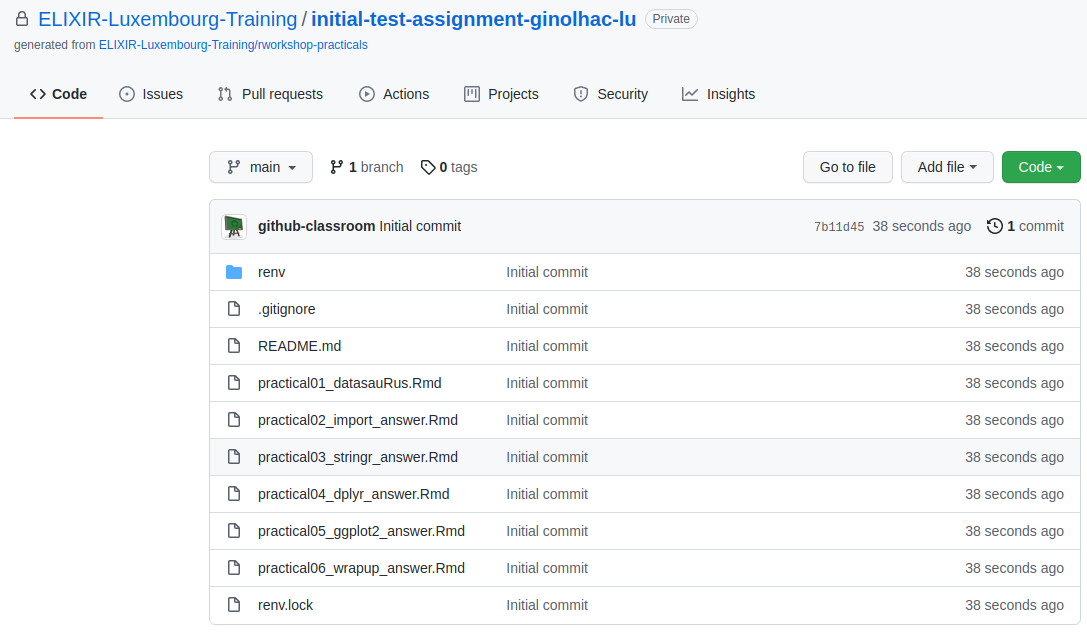
Copy the Code SSH URL
Make sure to use the Git Clone with SSH! 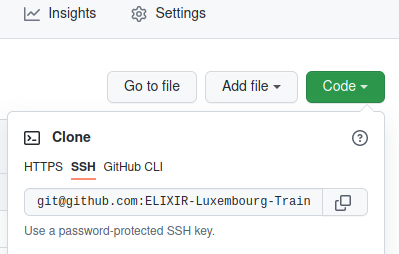
Clone the repository and complete the practical
Insert the URL for a new Git project
In Repository URL

Complete the actual assignment
Complete the instructions given in the README.md document.
Before we stop
You learned about:
- RStudio
- Basic functionality
- Recommended settings
- Project setup
- Code versioning with Git
- Simplify your analysis code
- GitHub classroom
- Package management with renv
Acknowledgments
- Hadley Wickham (RStudio)
- Kevin Ushey (
renv) - Romain François
- Jenny Bryan (File organization)
Contributors
- Roland Krause
- Aurélien Ginolhac
- Veronica Codoni
Thank you for your attention!