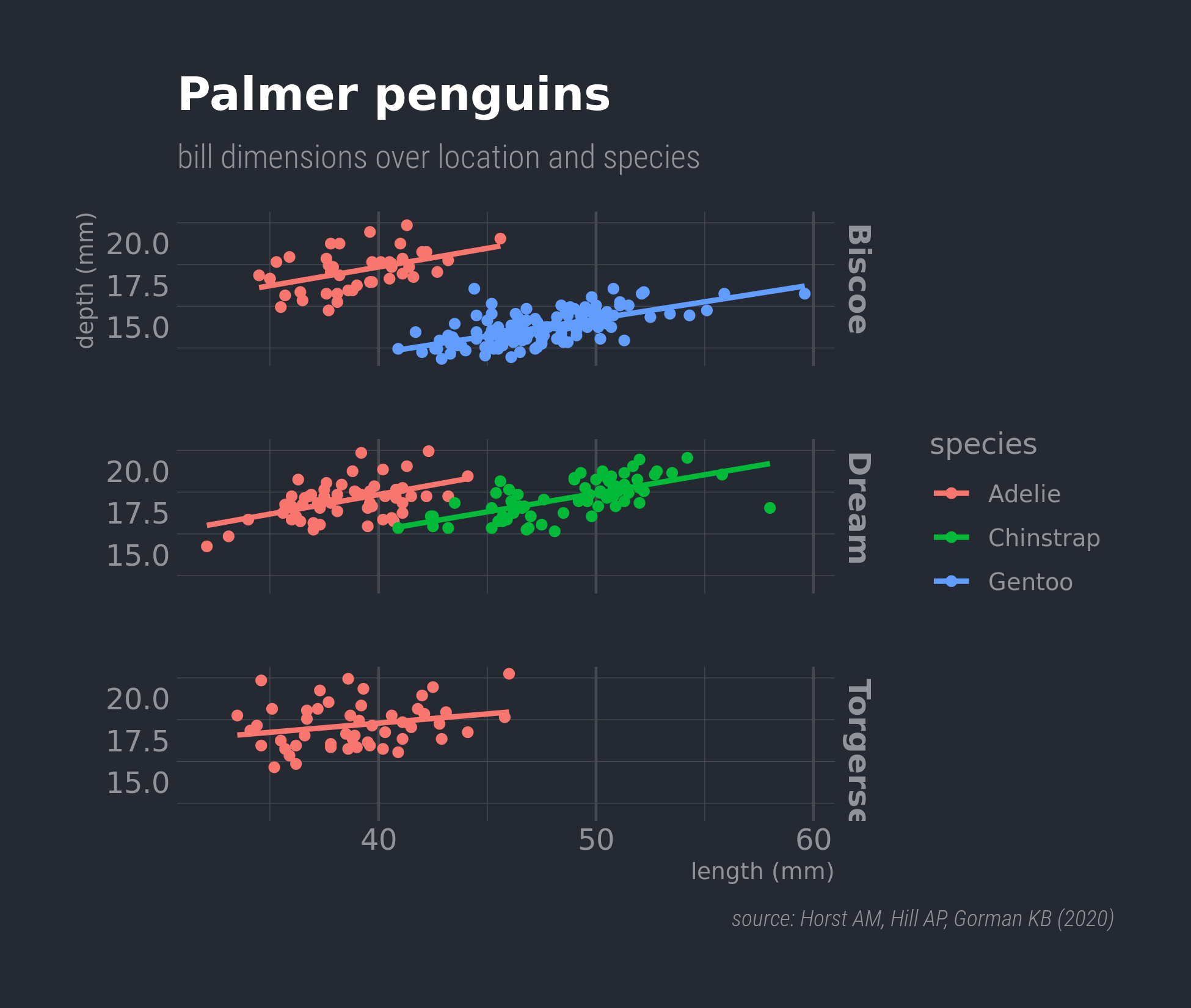
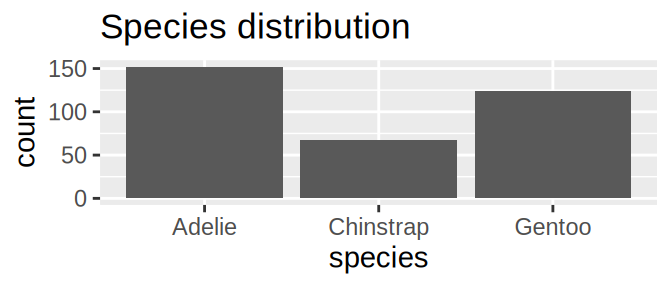
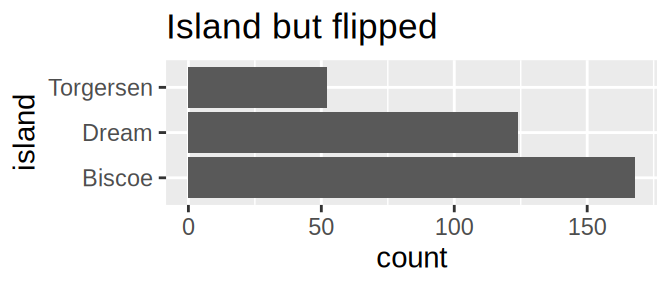
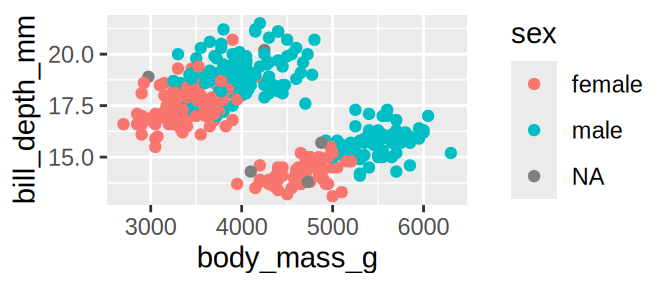
tidyr::crossing(aes = c("fill", "colour", "alpha", "x", "y"),
type = c("continuous", "discrete", "date")) |>
glue::glue_data("scale_{aes}_{type}()")scale_alpha_continuous()
scale_alpha_date()
scale_alpha_discrete()
scale_colour_continuous()
scale_colour_date()
scale_colour_discrete()
scale_fill_continuous()
scale_fill_date()
scale_fill_discrete()
scale_x_continuous()
scale_x_date()
scale_x_discrete()
scale_y_continuous()
scale_y_date()
scale_y_discrete()




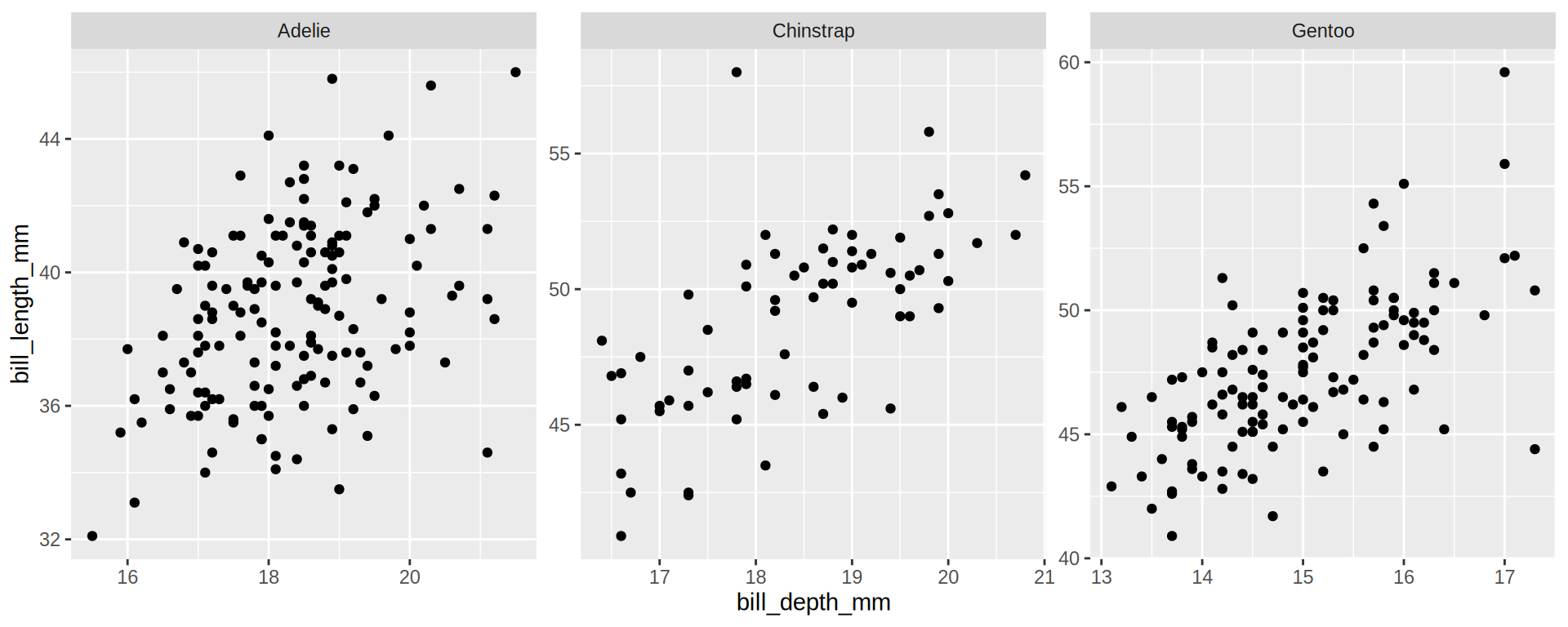
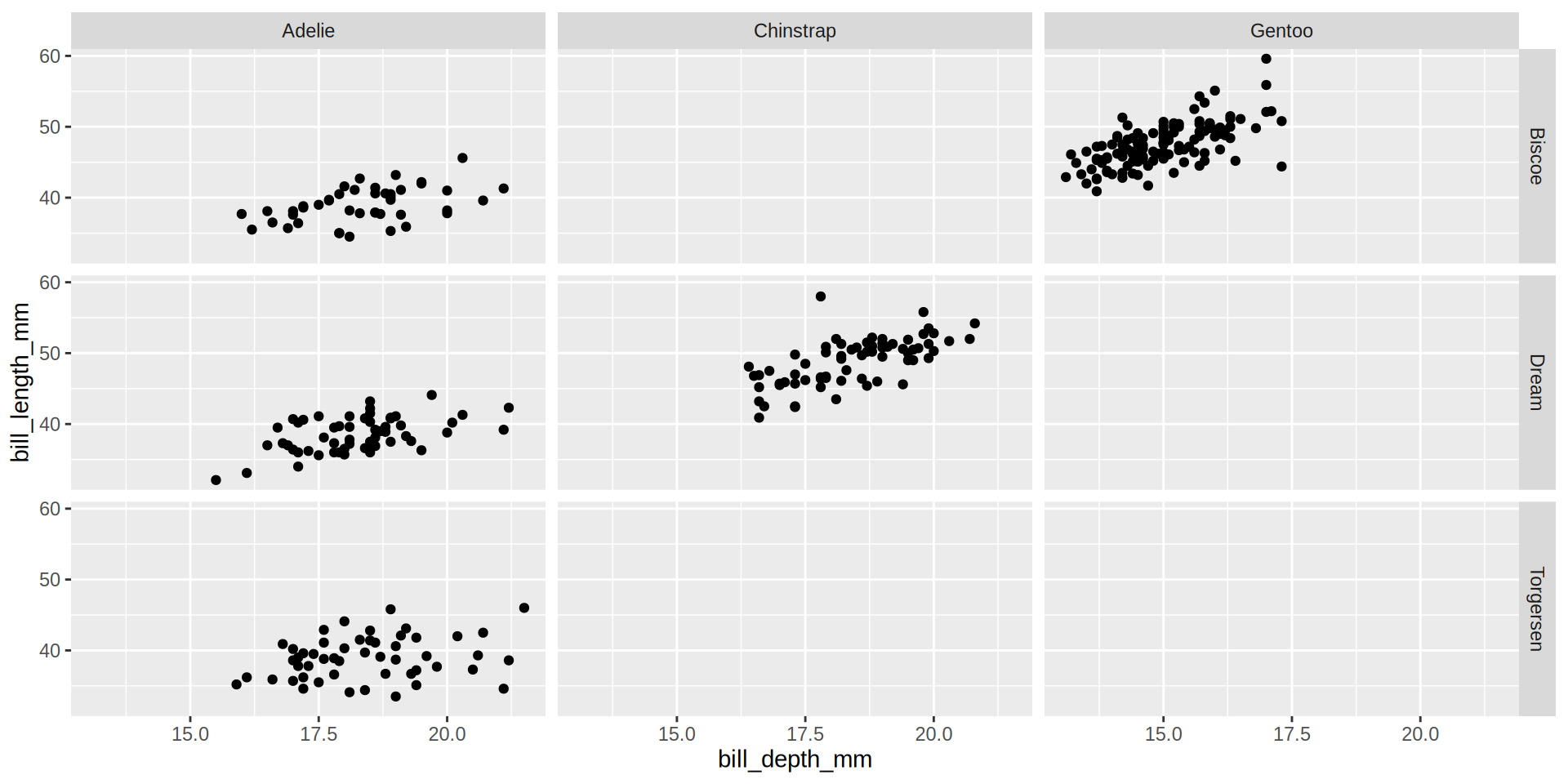
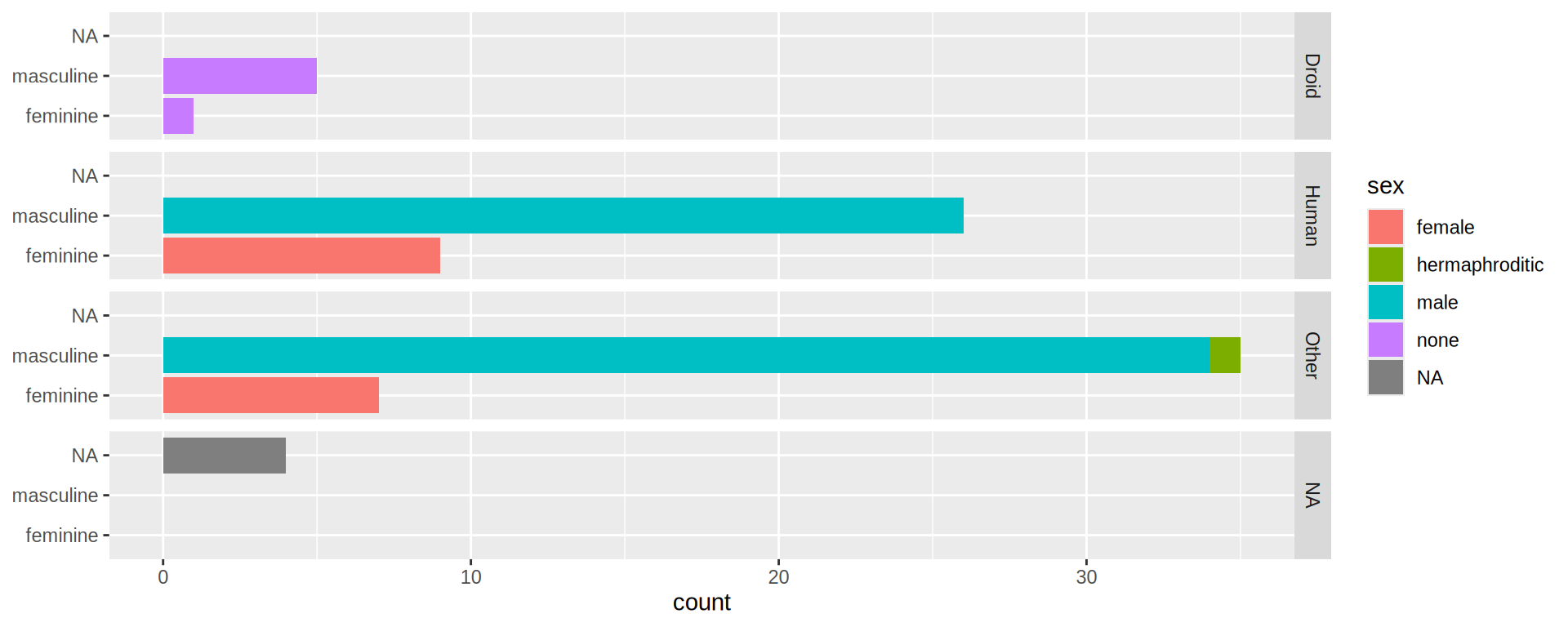
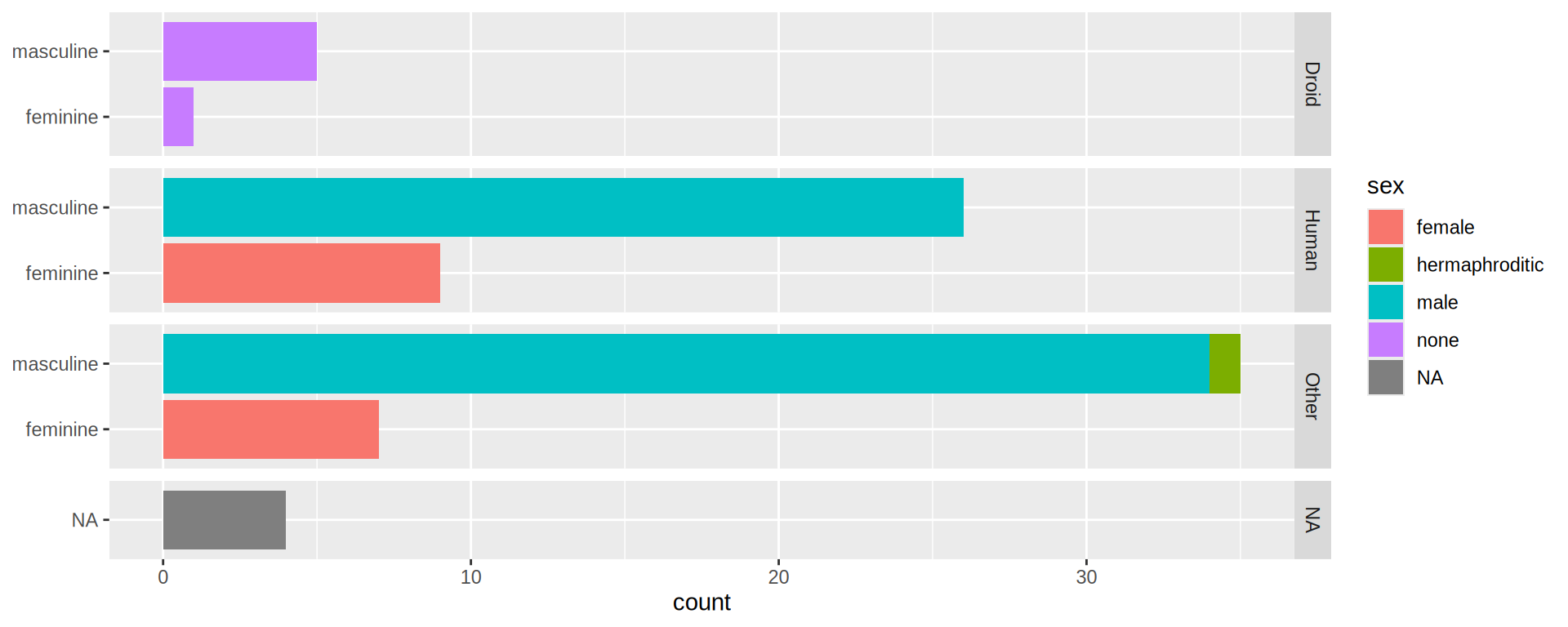
 (https://exts.ggplot2.tidyverse.org/gallery/)
(https://exts.ggplot2.tidyverse.org/gallery/)