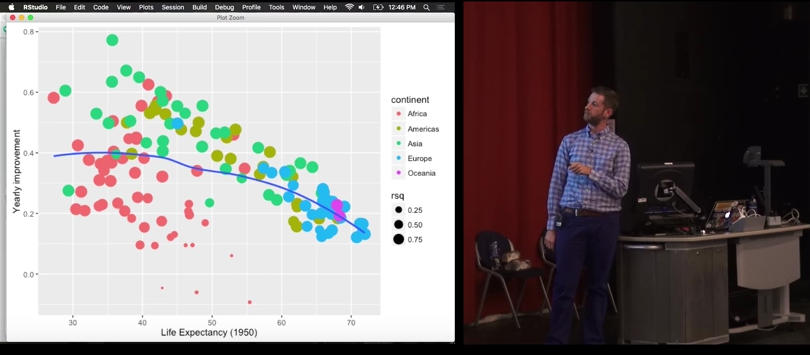
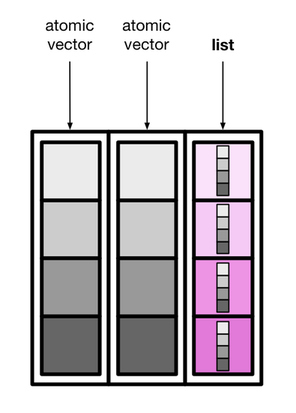
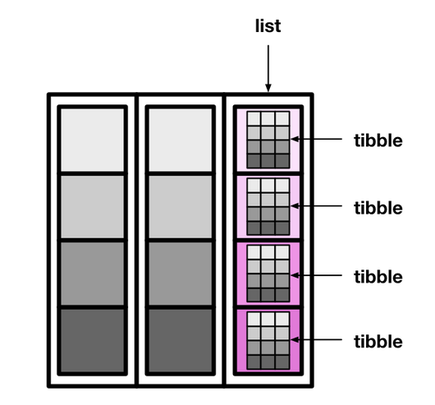
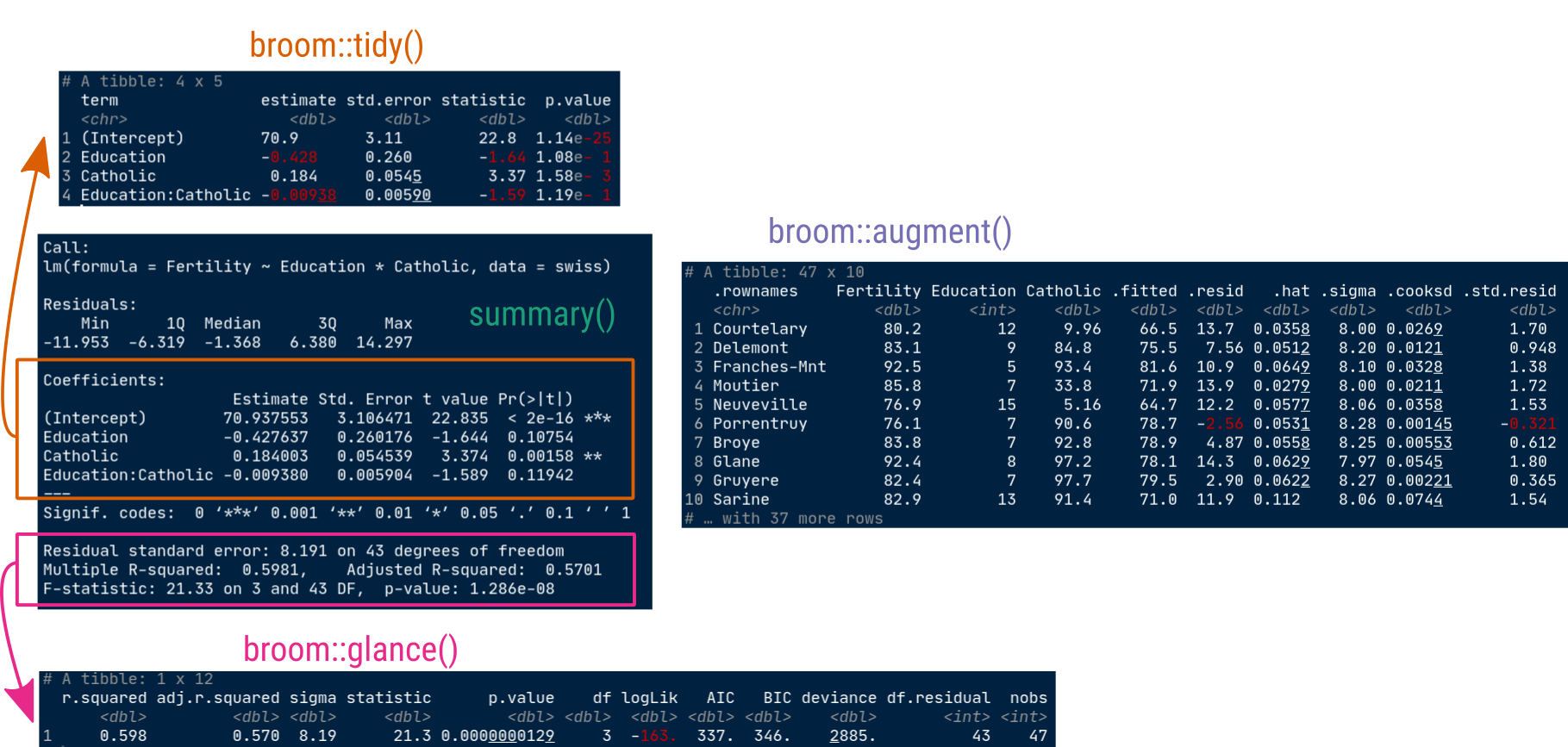
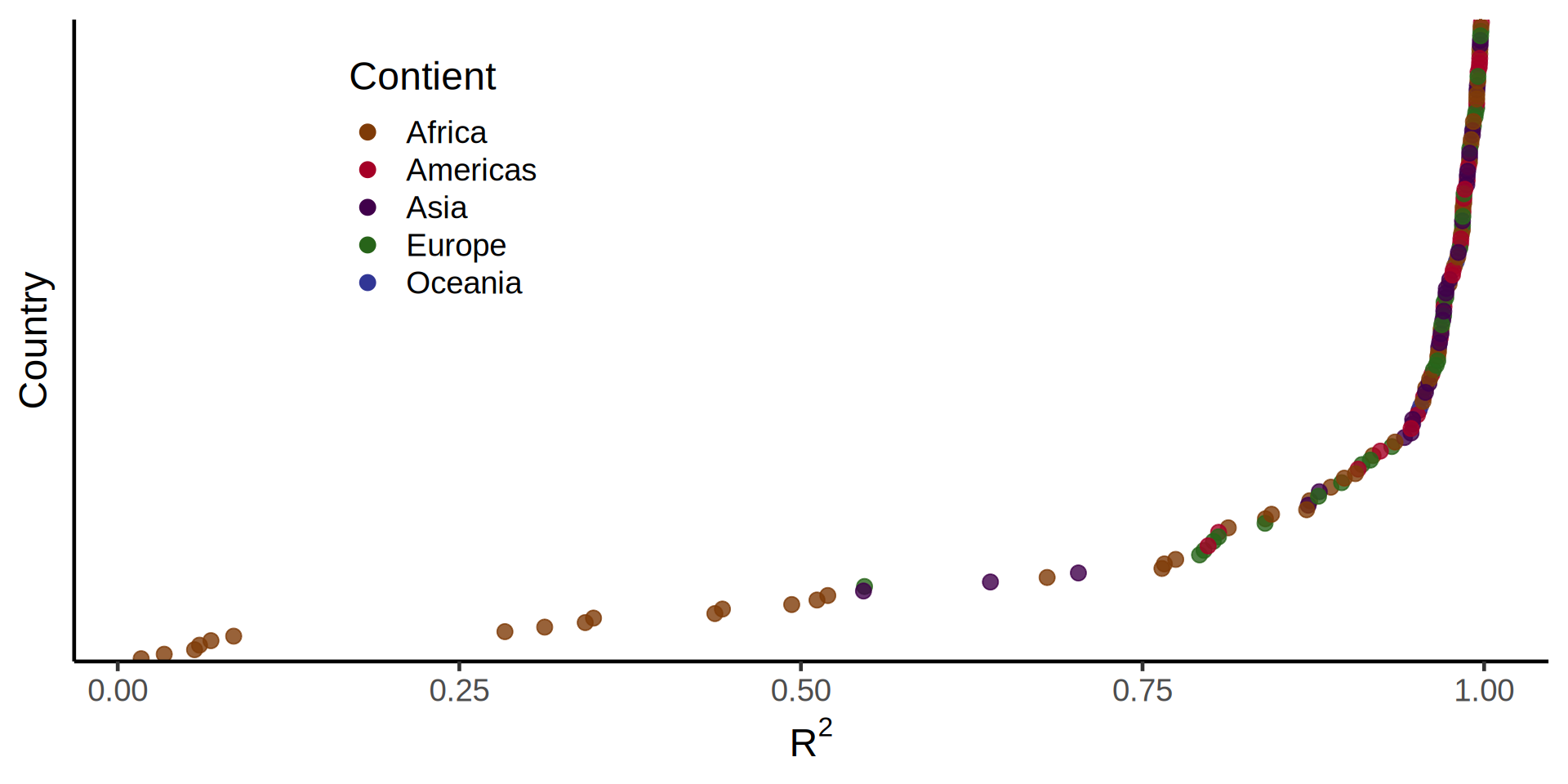
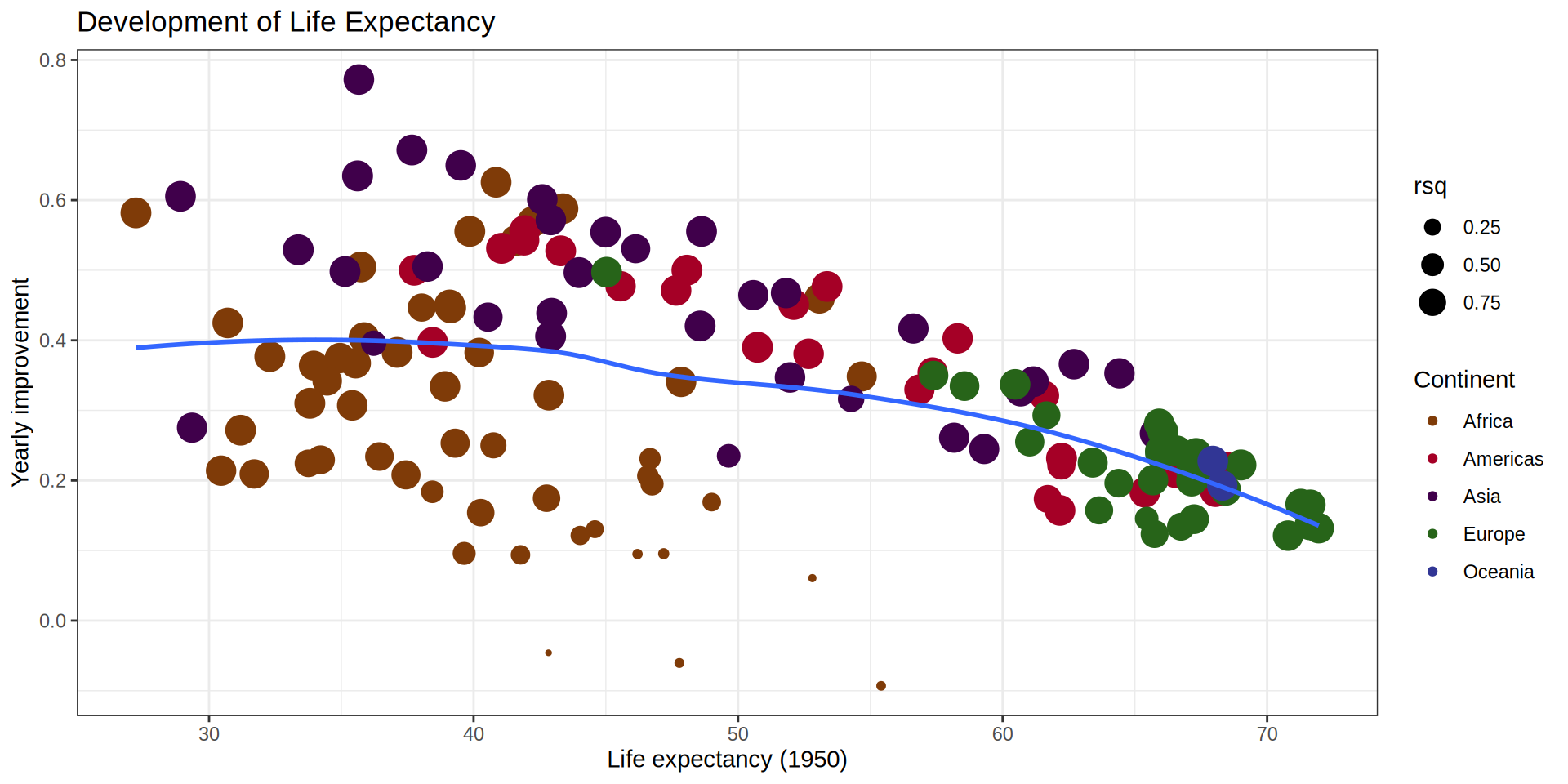
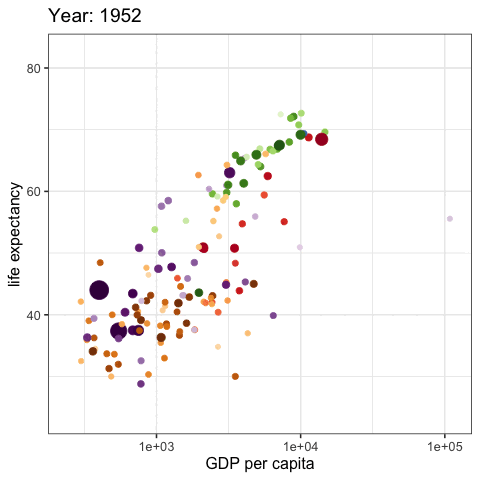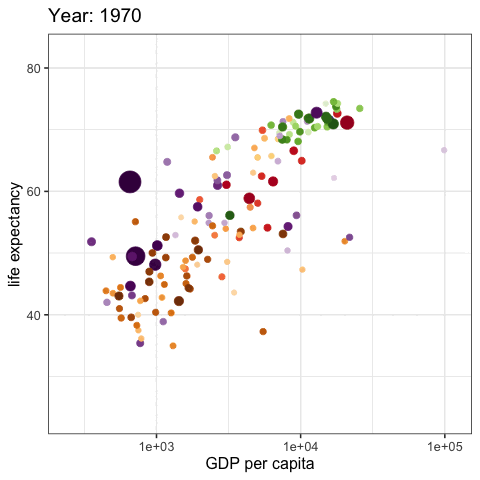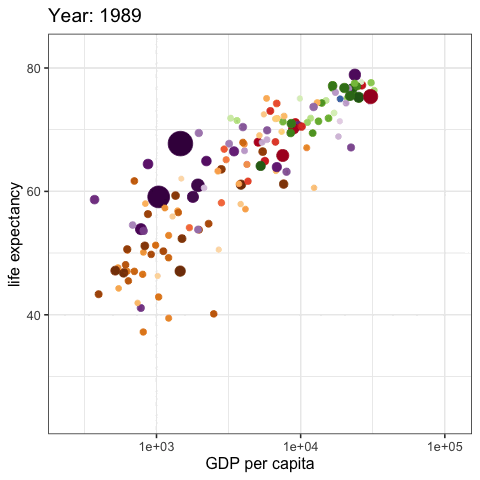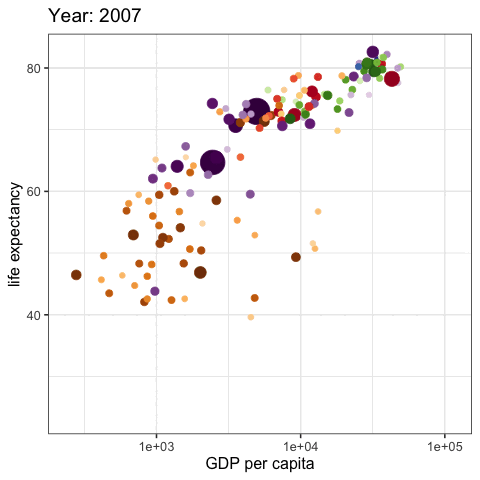
palmerpenguins::penguins |>
# Nest only the relevant cols (bill measurements)
# and ID penguins with biology criteria
nest(data = starts_with("bill"),
.by = c(island, species)) |>
mutate(model = map(data, \(x){ lm(bill_depth_mm ~ bill_length_mm,
data = x) } ),
summary = map(model, summary),
r_squared = map_dbl(summary, "r.squared"))# A tibble: 5 × 6
island species data model summary r_squared
<fct> <fct> <list> <list> <list> <dbl>
1 Torgersen Adelie <tibble [52 × 2]> <lm> <smmry.lm> 0.0620
2 Biscoe Adelie <tibble [44 × 2]> <lm> <smmry.lm> 0.219
3 Dream Adelie <tibble [56 × 2]> <lm> <smmry.lm> 0.258
4 Biscoe Gentoo <tibble [124 × 2]> <lm> <smmry.lm> 0.414
5 Dream Chinstrap <tibble [68 × 2]> <lm> <smmry.lm> 0.427