03:00
Joins and pivots
Merging data
Rworkshop
Tuesday, 11 February 2025
Introduction
Motivation
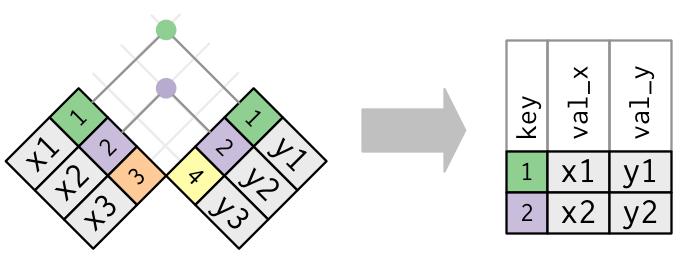
Joining data frames
How do we refer to a particular observation?
Primary key
The column that uniquely identifies an observation: - Frequently an ID - Can be introduced by row_number if no primary column exists bona fide observations

Composite key
Several columns that together identify an observation uniquely to serve as primary key.
Like patient and visit
Foreign key
A key referring to observations in another table
Your turn!
Tibbles do not have row names!
Frequently these are the primary keys! Use as_tibble(data, rownames = "ID") to make the rownames into a proper column.
Which columns are keys?
In
judgmentsIn
treesIn
swiss
Solution
Solution - Trees
Composite key from all the variables
Or a primary key from row numbers
Solution swiss
Province is primary key.
Once you took care of the rownames.
# A tibble: 47 × 7
Province Fertility Agriculture Examination Education
<chr> <dbl> <dbl> <int> <int>
1 Courtelary 80.2 17 15 12
2 Delemont 83.1 45.1 6 9
3 Franches-Mnt 92.5 39.7 5 5
4 Moutier 85.8 36.5 12 7
5 Neuveville 76.9 43.5 17 15
6 Porrentruy 76.1 35.3 9 7
7 Broye 83.8 70.2 16 7
8 Glane 92.4 67.8 14 8
9 Gruyere 82.4 53.3 12 7
10 Sarine 82.9 45.2 16 13
# ℹ 37 more rows
# ℹ 2 more variables: Catholic <dbl>,
# Infant.Mortality <dbl>Mutating joins
Matching subjects in two tibbles
Additional data on coffee consumption
Manual input using tribble()
Smaller sample set
From judgements
subject_mood <- judgments |>
select(subject, condition, gender,
starts_with("mood")) |>
distinct()
subject_mood# A tibble: 187 × 5
subject condition gender mood_pre mood_post
<dbl> <chr> <chr> <dbl> <dbl>
1 2 control female 81 NA
2 1 stress female 59 42
3 3 stress female 22 60
4 4 stress female 53 68
5 7 control female 48 NA
6 6 stress female 73 73
7 5 control female NA NA
8 9 control male 100 NA
9 16 stress female 67 74
10 13 stress female 30 68
# ℹ 177 more rowsThis is made up data for demonstration purposes only.
Combining the two tables
Inner join

Error! no common key.
Provide the corresponding columns names
Mutating joins
Inner join

Creating new tables through joins
Key operations in data processing
Role of observations as row changes
inner_join()is the most strict join operationsmergeis a similar operation in base R
Full Join
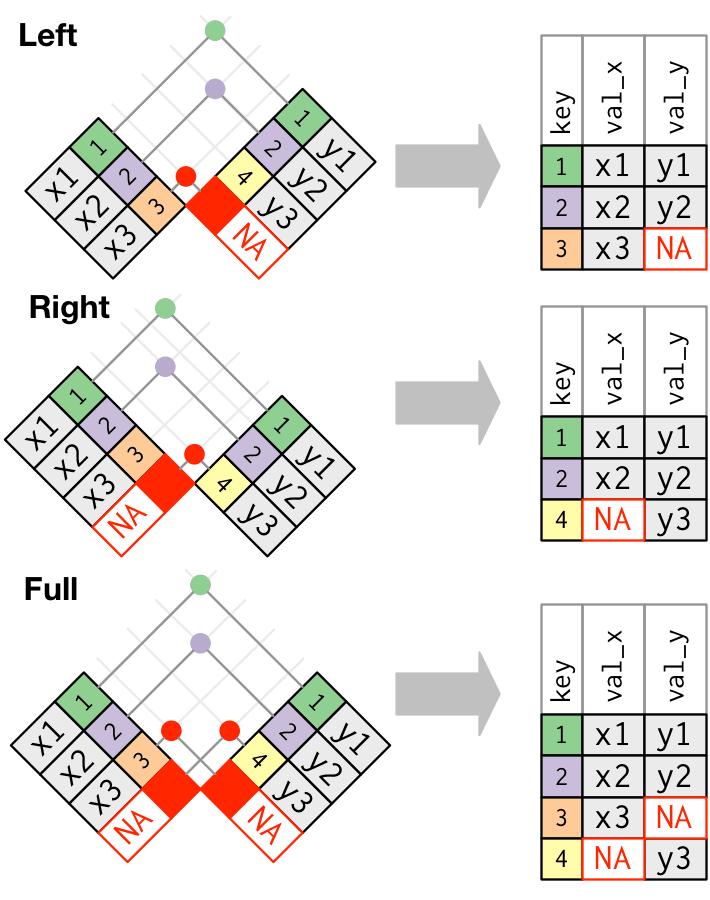
Example with full_join()
# A tibble: 187 × 6
subject condition gender mood_pre mood_post coffee_shots
<dbl> <chr> <chr> <dbl> <dbl> <dbl>
1 2 control female 81 NA NA
2 1 stress female 59 42 NA
3 3 stress female 22 60 NA
4 4 stress female 53 68 NA
5 7 control female 48 NA NA
6 6 stress female 73 73 NA
7 5 control female NA NA NA
8 9 control male 100 NA NA
9 16 stress female 67 74 NA
10 13 stress female 30 68 NA
# ℹ 177 more rowsTwo tables - same column names
What if we have gender in both tables?
# A tibble: 187 × 7
subject condition gender.x mood_pre mood_post
<dbl> <chr> <chr> <dbl> <dbl>
1 2 control female 81 NA
2 1 stress female 59 42
3 3 stress female 22 60
4 4 stress female 53 68
5 7 control female 48 NA
6 6 stress female 73 73
7 5 control female NA NA
8 9 control male 100 NA
9 16 stress female 67 74
10 13 stress female 30 68
# ℹ 177 more rows
# ℹ 2 more variables: coffee_shots <dbl>, gender.y <chr>Need to disambiguate columns
Mutating joins distinguish the same column names by adding suffixes - .x and .y by default.
Two tables - same column names
Join by one column
right_join(subject_mood,
coffee_drinkers,
by = join_by(subject == student),
suffix = c( "_mood", "_coffee"))# A tibble: 3 × 7
subject condition gender_mood mood_pre mood_post
<dbl> <chr> <chr> <dbl> <dbl>
1 23 control female 78 NA
2 21 control female 68 NA
3 28 stress male 53 68
# ℹ 2 more variables: coffee_shots <dbl>,
# gender_coffee <chr>Controlling suffix names
Add suffix as additional parameter
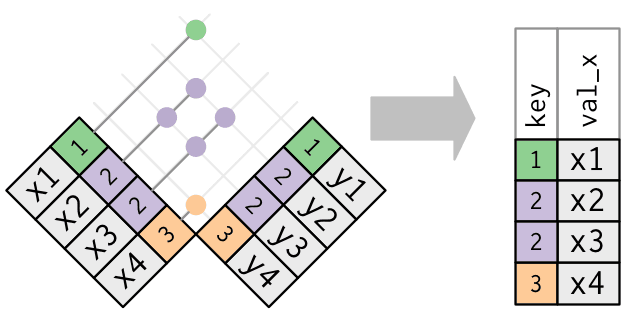
Filtering joins
Filtering joins -
semi_join()
Filter matches in x, no duplicates
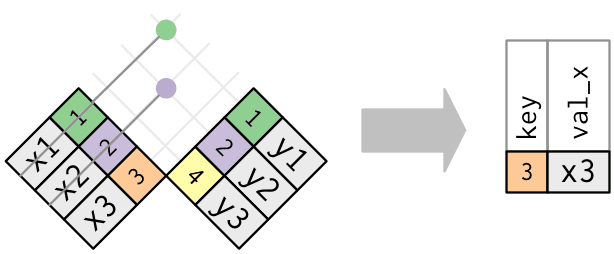
anti_join()
Extract what does not match
semi_join() to filter
Inequality joins
Inequality joins
Redefine matches keys
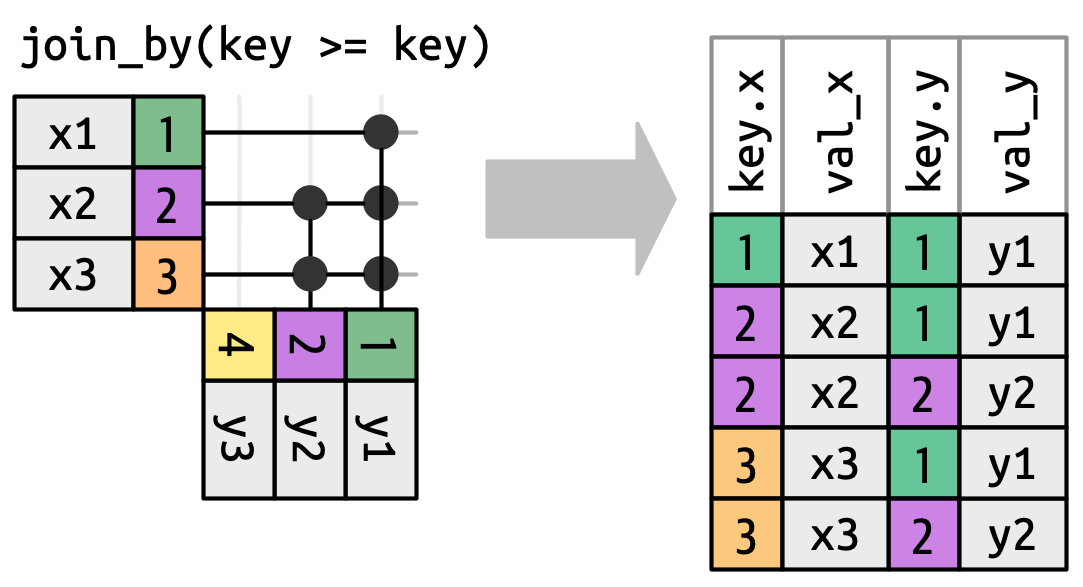
Caution
Unequal joins generate many additional rows.
Types of inqual joins
- Cross joins match every pair of rows.
- Inequality joins use
<,<=,>, and>=instead of==. - Rolling joins are similar to inequality joins but only find the closest match.
- Rolling joins allow to replace
if/elseorswitchstatements.
- Rolling joins allow to replace
- Overlap joins are a special type of inequality join designed to work with ranges.
Rewriting case_when()
Remember the tidyverse switch?
# A tibble: 188 × 3
subject mood_pre mood_pre_cat
<dbl> <dbl> <chr>
1 2 81 exceptional
2 1 59 great
3 3 22 poor
4 4 53 great
5 7 48 mid
6 6 73 great
7 5 NA missing data
8 9 100 exceptional
9 16 67 great
10 13 30 mid
# ℹ 178 more rowsRewriting case_when()
Remember the tidyverse switch?
# A tibble: 188 × 3
subject mood_pre mood_pre_cat
<dbl> <dbl> <chr>
1 2 81 exceptional
2 1 59 great
3 3 22 poor
4 4 53 great
5 7 48 mid
6 6 73 great
7 5 NA missing data
8 9 100 exceptional
9 16 67 great
10 13 30 mid
# ℹ 178 more rowsReplacement
Replace with a table and a join
Join
left_join(subject_mood, mood_categories,
by = join_by(mood_pre < score),
keep = TRUE) |>
select(subject, mood_pre, score, label)# A tibble: 394 × 4
subject mood_pre score label
<dbl> <dbl> <dbl> <chr>
1 2 81 100 exceptional
2 1 59 75 great
3 1 59 100 exceptional
4 3 22 25 poor
5 3 22 50 mid
6 3 22 75 great
7 3 22 100 exceptional
8 4 53 75 great
9 4 53 100 exceptional
10 7 48 50 mid
# ℹ 384 more rowsNot exactly helpful
Multiple entries (correctly) matching the condition.
Rolling join
Make it a rolling join with closest()
# A tibble: 187 × 4
subject mood_pre score label
<dbl> <dbl> <dbl> <chr>
1 2 81 100 exceptional
2 1 59 75 great
3 3 22 25 poor
4 4 53 75 great
5 7 48 50 mid
6 6 73 75 great
7 5 NA NA missing
8 9 100 100 exceptional
9 16 67 75 great
10 13 30 50 mid
# ℹ 177 more rowsReplacement
Replace with a table and a join
Join
left_join(subject_mood, mood_categories,
by = join_by(mood_pre < score),
keep = TRUE) |>
select(subject, mood_pre, score, label)# A tibble: 394 × 4
subject mood_pre score label
<dbl> <dbl> <dbl> <chr>
1 2 81 100 exceptional
2 1 59 75 great
3 1 59 100 exceptional
4 3 22 25 poor
5 3 22 50 mid
6 3 22 75 great
7 3 22 100 exceptional
8 4 53 75 great
9 4 53 100 exceptional
10 7 48 50 mid
# ℹ 384 more rowsNot exactly helpful
Multiple entries (correctly) matching the condition.
Rolling join
Make it a rolling join with closest()
# A tibble: 187 × 4
subject mood_pre score label
<dbl> <dbl> <dbl> <chr>
1 2 81 100 exceptional
2 1 59 75 great
3 3 22 25 poor
4 4 53 75 great
5 7 48 50 mid
6 6 73 75 great
7 5 NA NA missing
8 9 100 100 exceptional
9 16 67 75 great
10 13 30 50 mid
# ℹ 177 more rowsReshaping with pivot functions
Converting into long or wide formats - pivot functions
- The wide format is generally untidy but found in the majority of datasets and generally more readable.
- The long format makes computation on columns sometimes easier.
- Use
filterto compute on particular values.
- Use
- The functions are called
pivot_longer()andpivot_wider().
Pivot …
| site | 2001 | 2005 | 2015 |
|---|---|---|---|
| L1002 | 120 | 100 | 105 |
| L1034 | 125 | 130 | 140 |
| L1234 | 100 | 110 | 105 |
longer
| site | year | area |
|---|---|---|
| L1002 | 2001 | 120 |
| L1002 | 2005 | 100 |
| L1002 | 2015 | 105 |
| L1034 | 2001 | 125 |
| L1034 | 2005 | 130 |
| L1034 | 2015 | 140 |
| L1234 | 2001 | 100 |
| L1234 | 2005 | 110 |
| L1234 | 2015 | 105 |
Pivot …
| site | year | area |
|---|---|---|
| L1002 | 2001 | 120 |
| L1002 | 2005 | 100 |
| L1002 | 2015 | 105 |
| L1034 | 2001 | 125 |
| L1034 | 2005 | 130 |
| L1034 | 2015 | 140 |
| L1234 | 2001 | 100 |
| L1234 | 2005 | 110 |
| L1234 | 2015 | 105 |
wider
| site | 2001 | 2005 | 2015 |
|---|---|---|---|
| L1002 | 120 | 100 | 105 |
| L1034 | 125 | 130 | 140 |
| L1234 | 100 | 110 | 105 |
Making a wide data set longer
Calculations involving column and values
A toy data set
A longer table
area_long <-
pivot_longer(area_wide,
# columns to be transformed
cols = !contains("site"),
names_to = "year",
# We want to compute
names_transform = as.numeric,
values_to = "area")
area_long# A tibble: 9 × 3
site year area
<chr> <dbl> <dbl>
1 L1002 2001 120
2 L1002 2005 100
3 L1002 2015 105
4 L1034 2001 125
5 L1034 2005 130
6 L1034 2015 140
7 L1234 2001 100
8 L1234 2005 110
9 L1234 2015 105Making a wide data set longer
Calculations involving column and values
A toy data set
Variants by sample and positions.
A longer, tidier table
variants_long <-
pivot_longer(variants_wide,
cols = -contains("sample"), # columns argument, required
names_to = "pos",
values_to = "variant")
variants_long# A tibble: 9 × 3
sample_id pos variant
<chr> <chr> <chr>
1 L1002 3 A
2 L1002 5 C
3 L1002 8 <NA>
4 L1034 3 A
5 L1034 5 <NA>
6 L1034 8 T
7 L1234 3 <NA>
8 L1234 5 C
9 L1234 8 T Wider table
# A tibble: 188 × 10
subject condition age moral_dilemma_dog moral_dilemma_wallet moral_dilemma_plane moral_dilemma_resume
<dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2 control 24 9 9 8 7
2 1 stress 19 9 9 9 8
3 3 stress 19 8 7 8 5
4 4 stress 22 8 4 8 6
5 7 control 22 3 9 9 5
6 6 stress 22 9 9 9 9
7 5 control 18 9 5 7 3
8 9 control 20 9 4 1 7
9 16 stress 21 6 9 3 9
10 13 stress 19 6 8 9 8
# ℹ 178 more rows
# ℹ 3 more variables: moral_dilemma_kitten <dbl>, moral_dilemma_trolley <dbl>, moral_dilemma_control <dbl>Example
# A tibble: 1,316 × 5
subject condition age dilemma dilemma_val
<dbl> <chr> <dbl> <chr> <dbl>
1 2 control 24 dog 9
2 2 control 24 wallet 9
3 2 control 24 plane 8
4 2 control 24 resume 7
5 2 control 24 kitten 9
6 2 control 24 trolley 5
7 2 control 24 control 9
8 1 stress 19 dog 9
9 1 stress 19 wallet 9
10 1 stress 19 plane 9
# ℹ 1,306 more rowsExample
# A tibble: 18 × 3
condition dilemma_val n
<chr> <dbl> <int>
1 control 1 25
2 control 2 31
3 control 3 48
4 control 4 44
5 control 5 49
6 control 6 65
7 control 7 103
8 control 8 110
9 control 9 162
10 stress 1 13
11 stress 2 35
12 stress 3 32
13 stress 4 51
14 stress 5 56
15 stress 6 54
16 stress 7 103
17 stress 8 133
18 stress 9 202Example
judgments |>
select(subject, condition, age,
starts_with("moral_dilemma") ) |>
pivot_longer(starts_with("moral_dilemma") ,
names_to = "dilemma",
values_to = "dilemma_val",
names_pattern = "moral_dilemma_(.*)") |>
count(condition, dilemma_val) |>
pivot_wider(names_from = dilemma_val,
values_from = n,
names_prefix = "score")# A tibble: 2 × 10
condition score1 score2 score3 score4 score5 score6 score7 score8 score9
<chr> <int> <int> <int> <int> <int> <int> <int> <int> <int>
1 control 25 31 48 44 49 65 103 110 162
2 stress 13 35 32 51 56 54 103 133 202Helpful tools
Removing the data frame context through pull()
Remark
By default tidyverse operations that receive a tibble as input return a tibble.
Comparing to data in other rows
Leading and lagging rows
lead() for data in following row and lag() for data in the row above
Calculate differences between subjects
Let’s assume the subject IDs are in order of the tests being conducted. What is the difference to the previous subject for the “initial mood”?
judgments |>
select(subject, mood_pre) |>
arrange(subject) |>
mutate(prev_mood_pre = lag(mood_pre),
mood_diff = mood_pre - lag(mood_pre))# A tibble: 188 × 4
subject mood_pre prev_mood_pre mood_diff
<dbl> <dbl> <dbl> <dbl>
1 1 59 NA NA
2 2 81 59 22
3 3 22 81 -59
4 4 53 22 31
5 5 NA 53 NA
6 6 73 NA NA
7 7 48 73 -25
8 8 59 48 11
9 9 100 59 41
10 10 72 100 -28
# ℹ 178 more rowsThe other 20% of dplyr
Occasionally handy
- Assembly:
bind_rows,bind_cols - Windows function,
min_rank,dense_rank,cumsum. See vignette - Working with list-columns
multidplyrfor parallelized code
SQL mapping allows database access
dplyrcode can be translated into SQL and query databases online (usingdbplyr)- Different types of tabular data (dplyr SQL backend, databases)
Your turn!
Exercises
Take the results from the previous exercise available as judgments_condition_stats and bring the data into a more readable format.
- Make the table longer. After this step, your tibble should contain three columns:
- The condition
- the name of the moral dilemma and stats (in one column)
- the values of the stats by moral dilemma.
- Split the moral dilemma and stats column. You can use
separate_wider_delimormutateto create a column that contains the moral dilemma (dog, trolley, etc) and the stats (median, min, etc.). - Make the table wider again and move the stats to individual columns.
Your turn!
Exercises
The tribble contains changes of the sequence of a gene. The format in the input is the expected sequence (the reference allele), the position and the variant, commonly called alternative allele. In T6G, T is the reference allele, 6 is the position (along the gene) and G is the variant allele.
Clean the table of genetic variants such that all variants appear as a column labeled by their position.
Select relevant variants. In this table the variants are labeled according to their effect on stability of the gene product.
Identify the subjects in the table
variantsthat carry variants labeled as damaging invariant_significancetable. The final output should be vector of sample ids.
You can use several join flavours and the %in% operator to achieve the same result.
Bigger data
Go for data.table
- See this interesting thread about comparing
data.tableversusdplyr data.table, see introduction is very efficient but the syntax is not so easy.- Main advantage: inline replacement (tidyverse is frequently copying)
- As a summary: tl;dr data.table for speed, dplyr for readability and convenience Prashanth Sriram
- Hadley recommends that for data > 1-2 Gb, if speed is your main matter, go for
data.table dtplyris for learning thedata.tablefromdplyrAPI inputtidytableis actually usingdata.tablebut withdplyrsyntaxdata.tablemight be not useful for specialized applications with high volumes such as genomics.

Before we stop
You learned to:
- Joining and intersecting tibbles
- Differ filtering from mutation joins
- Reshaping column headers and variables
Further reading
Chapter 19 Joins
Acknowledgments
- Hadley Wickham
- Lionel Henry
- Romain François
- Lise Vaudor nice blog
- Allison Horst for the great ArtWork
- Alexandre Courtiol for cheatsheets
- Jenny Bryan
Contributions
- Milena Zizovic
- Aurélien Ginolhac
Thank you for your attention!
