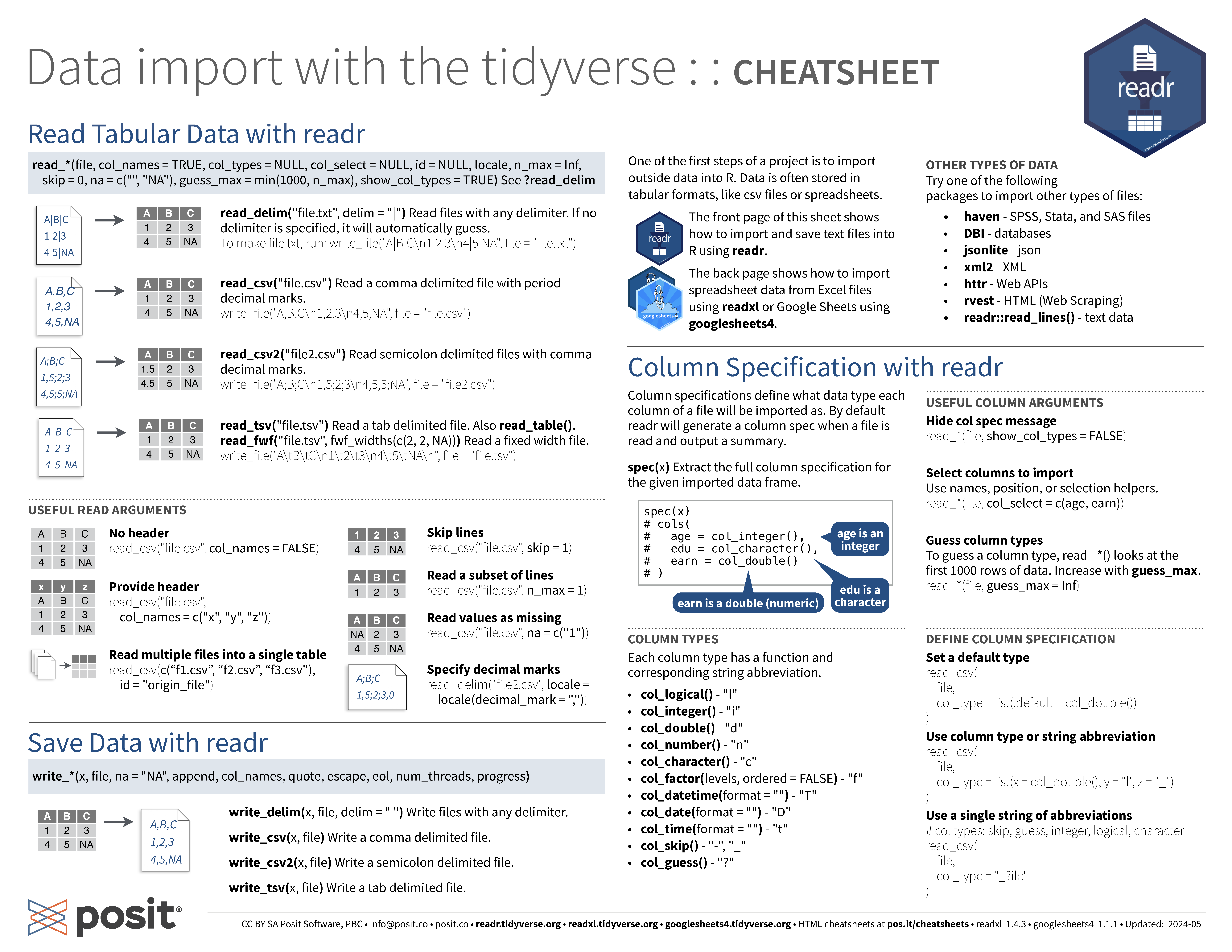
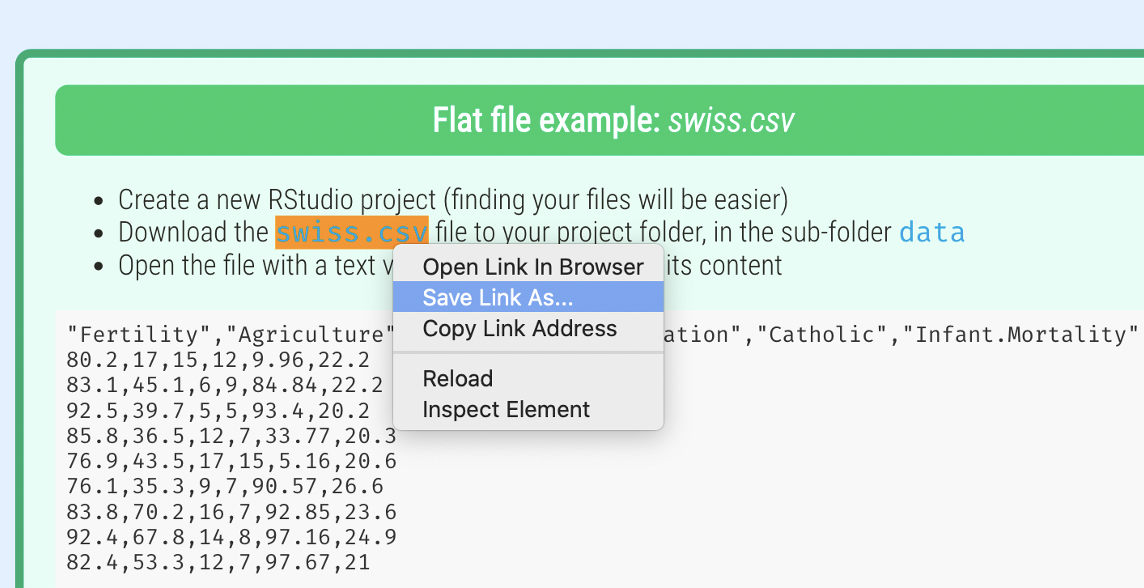
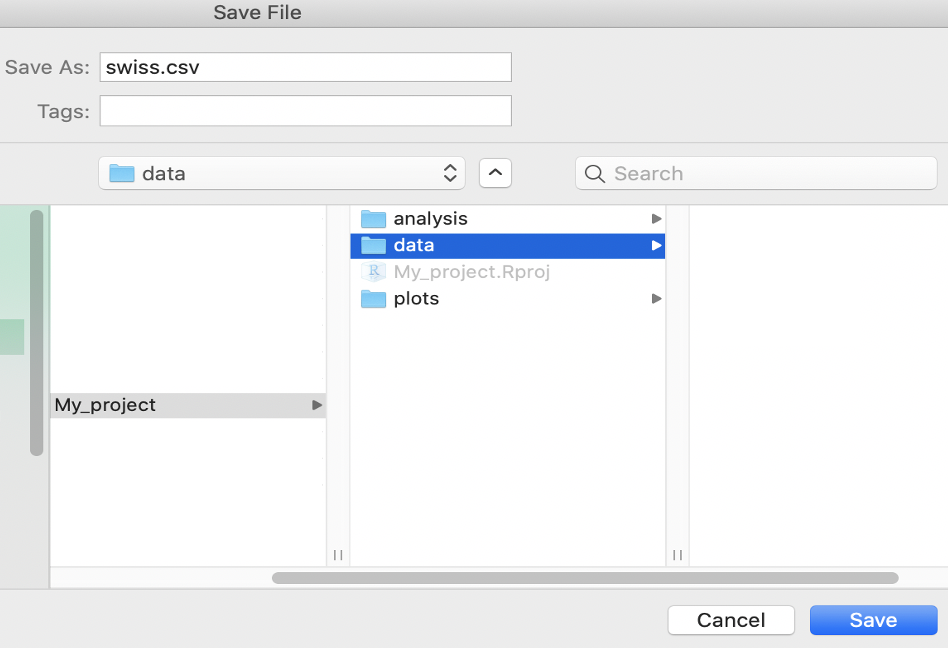
cols(
start_date = col_character(),
end_date = col_character(),
finished = col_double(),
condition = col_character(),
subject = col_double(),
gender = col_character(),
age = col_double(),
mood_pre = col_double(),
mood_post = col_double(),
STAI_pre_1_1 = col_double(),
STAI_pre_1_2 = col_double(),
STAI_pre_1_3 = col_double(),
STAI_pre_1_4 = col_double(),
STAI_pre_1_5 = col_double(),
STAI_pre_1_6 = col_double(),
STAI_pre_1_7 = col_double(),
STAI_pre_2_1 = col_double(),
STAI_pre_2_2 = col_double(),
STAI_pre_2_3 = col_double(),
STAI_pre_2_4 = col_double(),
STAI_pre_2_5 = col_double(),
STAI_pre_2_6 = col_double(),
STAI_pre_2_7 = col_double(),
STAI_pre_3_1 = col_double(),
STAI_pre_3_2 = col_double(),
STAI_pre_3_3 = col_double(),
STAI_pre_3_4 = col_double(),
STAI_pre_3_5 = col_double(),
STAI_pre_3_6 = col_double(),
STAI_post_1_1 = col_double(),
STAI_post_1_2 = col_double(),
STAI_post_1_3 = col_double(),
STAI_post_1_4 = col_double(),
STAI_post_1_5 = col_double(),
STAI_post_1_6 = col_double(),
STAI_post_1_7 = col_double(),
STAI_post_2_1 = col_double(),
STAI_post_2_2 = col_double(),
STAI_post_2_3 = col_double(),
STAI_post_2_4 = col_double(),
STAI_post_2_5 = col_double(),
STAI_post_2_6 = col_double(),
STAI_post_2_7 = col_double(),
STAI_post_3_1 = col_double(),
STAI_post_3_2 = col_double(),
STAI_post_3_3 = col_double(),
STAI_post_3_4 = col_double(),
STAI_post_3_5 = col_double(),
STAI_post_3_6 = col_double(),
moral_dilemma_dog = col_double(),
moral_dilemma_wallet = col_double(),
moral_dilemma_plane = col_double(),
moral_dilemma_resume = col_double(),
moral_dilemma_kitten = col_double(),
moral_dilemma_trolley = col_double(),
moral_dilemma_control = col_double(),
presentation_experience = col_double(),
presentation_unpleasant = col_double(),
presentation_fun = col_double(),
presentation_challenge = col_double(),
PBC_1 = col_double(),
PBC_2 = col_double(),
PBC_3 = col_double(),
PBC_4 = col_double(),
PBC_5 = col_double(),
REI_1 = col_double(),
REI_2 = col_double(),
REI_3 = col_double(),
REI_4 = col_double(),
REI_5 = col_double(),
REI_6 = col_double(),
REI_7 = col_double(),
REI_8 = col_double(),
REI_9 = col_double(),
REI_10 = col_double(),
REI_11 = col_double(),
REI_12 = col_double(),
REI_13 = col_double(),
REI_14 = col_double(),
REI_15 = col_double(),
REI_16 = col_double(),
REI_17 = col_double(),
REI_18 = col_double(),
REI_19 = col_double(),
REI_20 = col_double(),
REI_21 = col_double(),
REI_22 = col_double(),
REI_23 = col_double(),
REI_24 = col_double(),
REI_25 = col_double(),
REI_26 = col_double(),
REI_27 = col_double(),
REI_28 = col_double(),
REI_29 = col_double(),
REI_30 = col_double(),
REI_31 = col_double(),
REI_32 = col_double(),
REI_33 = col_double(),
REI_34 = col_double(),
REI_35 = col_double(),
REI_36 = col_double(),
REI_37 = col_double(),
REI_38 = col_double(),
REI_39 = col_double(),
REI_40 = col_double(),
MAIA_1_1 = col_double(),
MAIA_1_2 = col_double(),
MAIA_1_3 = col_double(),
MAIA_1_4 = col_double(),
MAIA_1_5 = col_double(),
MAIA_1_6 = col_double(),
MAIA_1_7 = col_double(),
MAIA_1_8 = col_double(),
MAIA_1_9 = col_double(),
MAIA_1_10 = col_double(),
MAIA_1_11 = col_double(),
MAIA_1_12 = col_double(),
MAIA_1_13 = col_double(),
MAIA_1_14 = col_double(),
MAIA_1_15 = col_double(),
MAIA_1_16 = col_double(),
MAIA_2_1 = col_double(),
MAIA_2_2 = col_double(),
MAIA_2_3 = col_double(),
MAIA_2_4 = col_double(),
MAIA_2_5 = col_double(),
MAIA_2_6 = col_double(),
MAIA_2_7 = col_double(),
MAIA_2_8 = col_double(),
MAIA_2_9 = col_double(),
MAIA_2_10 = col_double(),
MAIA_2_11 = col_double(),
MAIA_2_12 = col_double(),
MAIA_2_13 = col_double(),
MAIA_2_14 = col_double(),
MAIA_2_15 = col_double(),
MAIA_2_16 = col_double(),
STAI_pre = col_double(),
STAI_post = col_double(),
MAIA_noticing = col_double(),
MAIA_not_distracting = col_double(),
MAIA_not_worrying = col_double(),
MAIA_attention_regulation = col_double(),
MAIA_emotional_awareness = col_double(),
MAIA_self_regulation = col_double(),
MAIA_body_listening = col_double(),
MAIA_trusting = col_double(),
PBC = col_double(),
REI_rational_ability = col_double(),
REI_rational_engagement = col_double(),
REI_experiental_ability = col_double(),
REI_experiental_engagement = col_double(),
moral_judgment = col_double(),
moral_judgment_disgust = col_double(),
moral_judgment_non_disgust = col_double(),
presentation_evaluation = col_double(),
logbook = col_character(),
exclude = col_double()
)